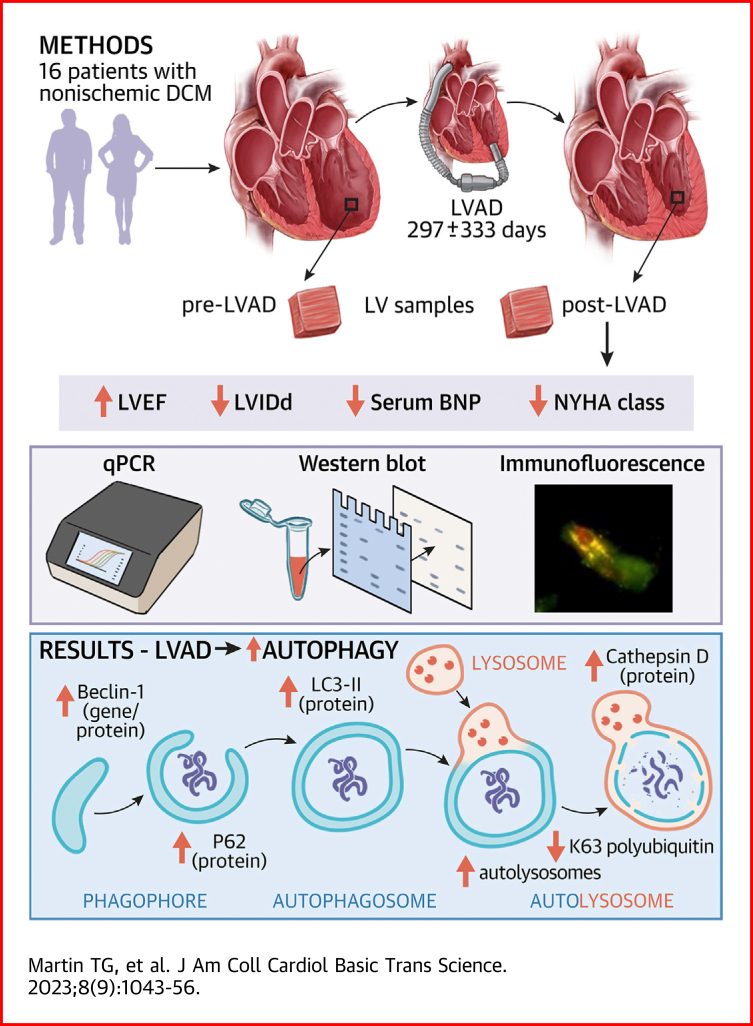
Visual Abstract
Key Words: autolysosome, autophagy, LVAD, reverse remodeling
Highlights
-
•
Paired left ventricle samples from 16 patients pre- and post-LVAD display improvements in pathological gene expression signatures post-LVAD.
-
•
Gene and protein expression of key autophagy factors increases following mechanical unloading by LVAD.
-
•
K63 polyubiquitinated proteins are reduced following LVAD, whereas autolysosome numbers increase, suggesting that a restoration of misfolded protein clearance by autophagy-lysosome pathway is restored with this therapy.
-
•
Enzymatic activity of calpains and the 20S proteasome increase post-LVAD and K48 polyubiquitinated proteins decrease, suggesting that proteolysis by these other 2 major protein degradation pathways also increases following mechanical unloading.
-
•
These results build on earlier studies indicating the involvement of autophagy in left ventricular reverse remodeling following mechanical unloading by LVAD and lend support for exploring the potential of autophagy-targeting therapies in heart disease.
Summary
Left ventricular reverse remodeling in heart failure is associated with improved clinical outcomes. However, the molecular features that drive this process are poorly defined. Left ventricular assist devices (LVADs) are the therapy associated with the greatest reverse remodeling and lead to partial myocardial recovery in most patients. In this study, we examined whether autophagy may be implicated in post-LVAD reverse remodeling. We found expression of key autophagy factors increased post-LVAD, while autophagic substrates decreased. Autolysosome numbers increased post-LVAD, further indicating increased autophagy. These findings support the conclusion that mechanical unloading activates autophagy, which may underly the reverse remodeling observed.
Heart disease is the leading cause of morbidity and mortality in the industrialized world. For many patients with end-stage heart failure, left ventricular assist devices (LVADs) are used either as a bridge to transplantation or as destination therapy.1 Numerous studies show that LVAD therapy is associated with reduced mortality and reverse left ventricular (LV) remodeling in the majority of patients.2 Hundreds of clinical studies over the past 3 decades indicate that regression of cardiac hypertrophy is associated with improved LV function in heart failure patients.3 The structural and functional improvements associated with LVAD, which stem from reduced LV wall stress, occur more rapidly and to a greater magnitude than with any other currently administered heart disease therapy.3 Thus, paired pre- and post-LVAD patient samples represent an extremely valuable resource for examining the molecular features of regression from pathological hypertrophy in humans.
Macroautophagy (hereafter, autophagy) is an evolutionarily conserved cellular recycling mechanism whereby long-lived proteins, protein aggregates, and whole organelles are encapsulated in a double-membraned autophagosome and delivered to a lysosome for degradation.4 Cardiomyocyte autophagy increases transiently in the initial response to pressure overload5; however, with chronic stimulation, it declines to levels below the normal steady state, contributing to heart failure.5,6 In multiple preclinical studies, mechanical unloading after a pathological stimulus resulted in increased autophagy, which was associated with regression of cardiomyocyte hypertrophy.7, 8, 9 Moreover, a recent clinical study of patients who exhibited reverse LV remodeling with first-line pharmacological heart disease therapies found that increased autophagy was predictive of this favorable outcome.10 In contrast, a study of 9 pre- and post-LVAD samples found expression of autophagy genes and the autophagosome protein LC3 decreased post-LVAD, leading the authors to conclude autophagy was reduced.11 The apparent incongruency in the conclusions of these studies is common in the heart failure literature on autophagy.12,13 In part, this is caused by different animal models, severity of cardiac stress, stage of heart failure, and disease etiology studied, all of which may affect autophagy.12,14 However, results from studies using LC3-II levels (ie, mature autophagosome number) as the sole readout for autophagy, without measuring additional markers of autophagy, have also contributed to the controversy around autophagy’s role in heart failure.14 Therefore, further study of autophagy following mechanical unloading by LVAD is warranted.
Here, we sought to examine markers of autophagy in 16 paired pre- and post-LVAD patient samples. Improved left ventricular ejection fraction (LVEF) post-LVAD displayed a significant positive correlation with regression of LV chamber diameter in these patients. Reduction of pathological gene expression signatures was also observed post-LVAD. Using quantitative polymerase chain reaction (qPCR), we found autophagy genes were differentially affected post-LVAD, with BECN1 and GABARAPL1 increasing and BNIP3 decreasing. We then assessed protein markers of autophagy and identified increased Beclin-1, LC3-II, P62, and cathepsin D expression, along with decreased K63-polyubiquitinated proteins suggestive of increased autophagic substrate removal post-LVAD. To further examine this process, we analyzed colocalization of autophagosomes with lysosomes (autolysosomes) by immunofluorescence microscopy and found a significant increase post-LVAD. Our findings support the conclusion that autophagy activity increases following LVAD therapy.
Methods
Tissue procurement and ethics approval
Paired LV tissue samples were collected from 16 patients with end-stage heart failure undergoing bridge to transplant LVAD implantation (pre-LVAD) and then subsequently at the time of cardiac transplantation (post-LVAD). The samples were immediately flash frozen in liquid nitrogen in the operating room and then stored at −80 °C. Corresponding deidentified clinical data were recorded for each patient in a secure REDCap database. The Colorado Multiple Institutional Review Board approved the protocol for collection, storage, and analysis of human tissue.
RNA extraction, cDNA synthesis, and qPCR analysis
LV tissue was homogenized in 1 mL Trizol, transferred to 1.5-mL Eppendorf tubes, and incubated at room temperature for 5 minutes. Then, 200 μL chloroform was added, the tubes were shaken vigorously, and they were incubated at room temperature for 15 minutes. Following this incubation, the samples were centrifuged at 12,000 RCF and 4 °C for 15 minutes. The top aqueous layer was transferred to a new tube, combined with an equal volume of isopropanol, and incubated at room temperature for 10 minutes. The samples were centrifuged at 12,000 RCF for 8 minutes and the supernatant discarded. The RNA pellet was washed with ice-cold 75% ethanol and then solubilized in sterile milli-Q H2O. A total of 500 ng of RNA was used for cDNA synthesis with random primers and Superscript III reverse transcriptase. Quantitative PCR was performed with 5 ng cDNA input using SYBR Green with the standard curve method. Primer sequences are available in Supplemental Table 1. Expression of genes of interest was normalized to the housekeeping gene 18S. Outliers >2 SDs from the mean were removed before analysis.
Protein extraction and Western blot analysis
Frozen LV tissue samples were pulverized and added to RIPA buffer containing protease and phosphatase inhibitors (Thermo Fisher, 78442). Samples were incubated with constant rotation overnight at 4 °C. They were then centrifuged at 12,000 RCF for 10 minutes at 4 °C, and the supernatant was transferred to a clean tube. Protein concentration was determined by BCA assay (Pierce). Lysates were then diluted to 1 μg/μL in gel loading buffer (3:1 Tris-Glycine SDS Sample Buffer [Novex], Bolt Reducing Buffer [Novex]), boiled, and 10 μL loaded onto 4% to 12% gradient NuPAGE Bis-Tris gels (Invitrogen). Proteins were separated by electrophoresis and transferred onto nitrocellulose membrane. Revert Total Protein Stain (LI-COR) was used to confirm effective transfer. The membranes were blocked for 1 hour at room temperature in 5% milk. Primary antibodies were then applied at 1:1,000 dilution in 5% (w/v) bovine serum albumin fraction V (Millipore Sigma) in 0.2% tween tris-buffered saline and incubated overnight at 4 °C. Primary antibodies: VPS34 (Proteintech, 12452-1-AP), Beclin-1 (Cell Signaling, 3495), ATG5 (Cell Signaling, 12994), Gabarapl1 (Proteintech, 11010-1-AP), LC3B (Cell Signaling, 2775), P62/Sqstm1 (Cell Signaling, 5114), BNIP3 (Cell Signaling, 44060), Parkin (Proteintech, 14060-1-AP), PINK1 (Cell Signaling, 6946), Cathepsin D (Novus Biologicals, NBP1-04278), Lamp1 (Cell Signaling, 9091), Pan-ubiquitin (Cytoskeleton Inc, AUB01), K48 ubiquitin (Cell Signaling, 4289), K63 ubiquitin (Cell Signaling, 5621), Atrogin-1 (Proteintech, 67172-1-Ig), MuRF-1 (Proteintech, 55456-1-AP), and GAPDH (Cell Signaling, 2118). Secondary antibodies for the ubiquitin blots were LICOR IRdye 800cw antimouse or antirabbit immunoglobulin G (IgG) and were used at 1:5,000 dilution in blocking buffer for 1 hour at room temperature. Secondary antibodies for the remainder were goat antirabbit IgG HRP-linked (Cell Signaling 7074) or goat antimouse IgG HRP-linked (Cell Signaling 7076) and were used at a 1:2,000 dilution. Membranes were washed with tris-buffered saline, incubated briefly in ECL substrate, and imaged on an Azure c600 or ImageQuant LAS 4000 Biomolecular Imager. Densitometry was measured using LICOR Image Studio or ImageQuant TL.
Immunofluorescence microscopy
The protocol was adapted from that described previously.15 Frozen LV tissue was homogenized in 1 mL of isolation buffer (2 mmol/L EGTA, 8.9 mmol/L potassium hydroxide, 7.1 mmol/L MgCl2, 5.8 mmol/L ATP, 10 mmol/L imidazole, 108 mmol/L potassium chloride) containing protease and phosphatase inhibitors, added to Chambered Coverglass (Nunc, Lab-Tek) precoated with poly-D-lysine, and incubated for 30 minutes at room temperature. The wells were washed once with phosphate buffered saline (PBS), and 0.5% triton X-100 was added and incubated for 20 minutes, followed by 2 10-minute incubations with 0.1% triton X-100. Antigen retrieval was performed by adding 100 mmol/L glycine (pH 7.4) for 30 minutes at room temperature. Wells were then washed 3× with PBS and blocking performed with 5% bovine serum albumin fraction V (in PBS) for 1 hour at room temperature. Primary antibodies for LC3B (Rabbit, Cell Signaling, 2775—1:100 dilution) and cathepsin D (Mouse, Novus Biologicals, NBP1-04278—1:100 dilution) were added in blocking solution and incubated overnight at 4 °C. On day 2, the wells were washed with PBS, and Alexa-Fluor secondary antibodies (Thermo Fisher, Anti-Rb 488, Anti-MS 568) were added in blocking solution at 1:1,000 dilution and incubated for 1 hour at room temperature. The wells were washed with PBS and slides were mounted in Vectashield with DAPI (Vectorlabs). Slides were imaged on a Nikon Widefield epi-fluorescence microscope using the 40× air objective. In total, 8 pre-LVAD and 8 post-LVAD samples were analyzed. A total of 10 to 12 images per sample were acquired at random, and the number of total lysosomes (cathepsin D positive puncta) and autolysosomes (LC3B/cathepsin D-positive puncta) were counted by an experimenter blinded to the experimental groups.
20S proteasome activity assay
Frozen tissue was pulverized and added to assay buffer (50 mmol/L HEPES pH 7.8, 10 mmol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EDTA, 1 mmol/L EGTA, 250 mmol/L sucrose). To allow for effective lysis, the samples were incubated for 3 hours at 4 °C with constant rotation. Lysates were centrifuged at 16,000 RCF for 15 minutes and the supernatants collected. The BCA assay was used to determine protein concentration. DTT was then added at a final concentration of 5 mmol/L, and 50 μL of 0.4 μg/μL sample was added in duplicate to a black-walled, clear-bottomed 96-well plate. The 50 μL assay buffer alone was used as a no protein control. Suc-LLVY-AMC substrate and ATP in 200 μL of assay buffer was then added to each well at final concentrations of 100 μmol/L and 2 mmol/L, respectively. For a negative control, the proteasome inhibitor Bortezomib was included at 20 μmol/L final concentration. The plate was incubated for 3 hours at 37 °C. Free AMC, cleaved from the peptide by the 20S proteasome, was measured on a plate reader at 390/460 nm. Data analysis was performed by subtracting the raw fluorescence values with Bortezomib and normalizing to the pre-LVAD group.
Calpain activity assay
Lysates were prepared as described in the previous text. DTT and CaCl2 were then added at final concentrations of 5 and 3 mmol/L, respectively. A total of 100 μL of 0.5 μg/μl sample was added in duplicate to a black-walled, clear-bottomed 96-well plate. Buffer alone was loaded as the no protein control. In total, 100 μL of assay buffer containing the calpain 1 and 2 substrate Ac-LLY-AFC was then added to the wells with a final substrate concentration of 200 μmol/L. Buffer and 200 μmol/L substrate without protein added served as the negative control. The plate was incubated at 37 °C for 1 hour. Free AFC, cleaved from the peptide by calpains 1 and 2, was measured on a plate reader at 400/505 nm. Data analysis was performed by subtracting the raw fluorescence of the negative control and normalizing to the pre-LVAD group.
Data presentation and statistical analysis
Data analysis and presentation were performed using GraphPad Prism version 9. Comparisons between pre- and post-LVAD were performed by paired 2-tailed Student’s t-test and comparisons of 2 independent groups using 2-tailed Student’s t-test, unless otherwise noted. Comparisons involving more than 2 groups were done by 1-way analysis of variance. When a significant interaction was identified, Tukey’s post hoc test for multiple pairwise comparisons was employed. Data containing 2 continuous variables were analyzed by linear regression analysis. Categorical data from pre- and post-LVAD samples were analyzed by McNemar’s test and are presented as count or percentage. Throughout the paper, the data are presented as the mean ± SEM unless otherwise noted. A P value <0.05 was considered significant.
Results
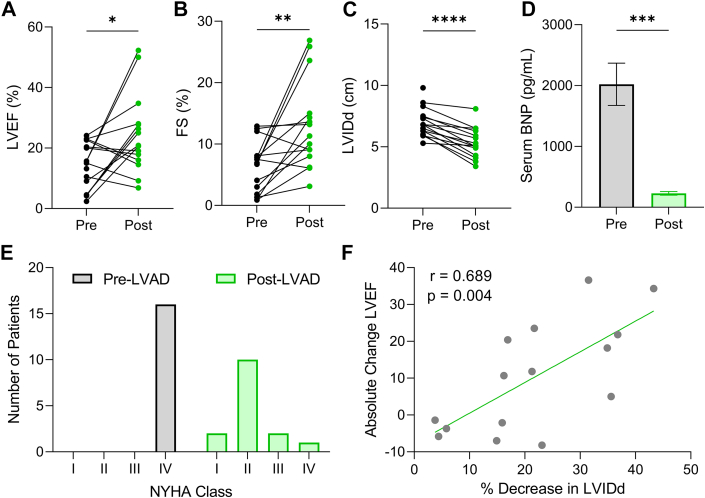
Paired pre- and post-LVAD samples (Table 1) were used to examine the impact of LVAD on LV hypertrophy regression and autophagy. All patients had nonischemic dilated cardiomyopathy, and at the time of LVAD implantation, all were characterized as having NYHA functional class IV heart failure (Table 2). As previously identified, LVAD therapy was associated with a significant improvement of LVEF (Table 1, Figure 1A) and fractional shortening (Table 1, Figure 1B) and a reduction in LV internal diameter (Table 1, Figure 1C). Serum B-type natriuretic peptide levels, a common indicator of cardiac pathology, also decreased post-LVAD (Figure 1D). Although all patients started in end-stage heart failure, following LVAD therapy, the majority (73.3%) improved to NYHA functional class I or II (Figure 1E). As expected, the reduction of LV chamber diameter positively correlated with the extent of myocardial functional recovery denoted by LVEF (Figure 1F). These clinical data reflect the recordings taken at the time nearest to tissue collection.
Table 1.
Patient Population
| Age at LVAD placement, y | 47.3 ± 13.3 |
| Male | 75.0 |
| White | 87.5 |
| Duration of LVAD support, d | 298.5 ± 333.1 |
Values are mean ± SD or %.
LVAD = left ventricular assist device.
Table 2.
Clinical Characteristics Pre- and Post-LVAD
| Pre-LVAD | Post-LVAD | P Value | |
|---|---|---|---|
| NYHA functional class, I/II/III/IV | 0/0/0/16 | 2/10/2/1a | <0.001 |
| LVEF, % | 14.2 ± 7.6 | 24.6 ± 13.0a | 0.019 |
| Fractional shortening, % | 6.3 ± 4.0 | 13.0 ± 7.3a | 0.003 |
| LVIDs, cm | 6.5 ± 1.2 | 4.7 ± 1.3a | <0.001 |
| LVIDd, cm | 7.0 ± 1.2 | 5.3 ± 1.2a | <0.001 |
| Heart rate, beats/min | 97.6 ± 20.9 | 88.9 ± 16.5 | 0.14 |
| Mean arterial pressure, mm Hg | 75.6 ± 11.4 | 82.3 ± 7.3 | 0.15 |
| Body weight, kg | 82.8 ± 17.8 | 81.7 ± 16.4 | 0.51 |
| Body surface area, m2 | 1.99 ± 0.23 | 1.98 ± 0.22 | 0.33 |
| Serum B-type natriuretic peptide, pg/mL | 1,765.6 ± 1,283.0 | 387.1 ± 599.6b | 0.002 |
| Serum total cholesterol, mg/dL | 121.1 ± 34.2 | 158.6 ± 28.2 | 0.003 |
| Serum creatinine, mg/dL | 1.27 ± 0.24 | 0.98 ± 0.21 | 0.001 |
| Medications | |||
| ACE inhibitor | 1 (6.3) | 9 (56.3) | 0.013 |
| Aldosterone antagonist | 14 (87.5) | 16 (100) | 0.48 |
| Antiarrhythmic agent | 4 (25.0) | 4 (25.0) | 0.62 |
| Beta-blocker | 2 (12.5) | 11 (68.8) | 0.008 |
| Digoxin | 8 (50.0) | 5 (31.3) | 0.45 |
| Dobutamine | 13 (81.3) | 0 (0.0) | <0.001 |
| Hydralazine | 3 (18.8) | 3 (18.8) | 0.68 |
| Loop diuretic | 14 (87.5) | 9 (56.3) | 0.13 |
| Milrinone | 2 (12.5) | 1 (6.3) | >0.99 |
| PDE5 inhibitor | 0 (0.0) | 1 (6.3) | >0.99 |
| Statin | 0 (0.0) | 2 (12.5) | 0.48 |
| Warfarin | 0 (0.0) | 14 (87.5) | <0.001 |
Values are mean ± SD or n (%). For medications, data are listed as n (%). Italic values indicate statistical significant.
ACE = angiotensin-converting enzyme; LVAD = left ventricular assist device; LVEF = left ventricular ejection fraction; LVIDd = left ventricular internal diameter during diastole; LVIDs = left ventricular internal diameter during systole.
Not measured in 1 patient.
Not measured in 7 patients. Data analysis by McNemar’s test for paired categorical data for NYHA functional class and medications. Data analysis by 2-tailed paired Student's t-test for all other comparisons.
Figure 1.
LVAD Therapy Is Associated With Reverse Remodeling and Partial Myocardial Recovery
(A) Left ventricular ejection fraction (LVEF) pre- and post-left ventricular assist device (LVAD). (B) Left ventricular fractional shortening (FS) pre- and post-LVAD. (C) Left ventricle internal diameter during diastole (LVIDd) pre- and post-LVAD; for A to C, n = 16 pre-LVAD, 15 post-LVAD (echocardiography data was not available for 1 patient post-LVAD). (D) Serum B-type natriuretic peptide levels pre- and post-LVAD; n = 16 pre-LVAD, 11 post-LVAD (not available for 7 patients); mean ± SEM. (E) NYHA functional class distribution pre- and post-LVAD; not available for 1 post-LVAD patient. (F) Absolute change in LVEF (recovery) and % decrease in LVIDd (regression) analyzed by linear regression. For A to D, data were analyzed by 2-tailed paired Student's t-test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
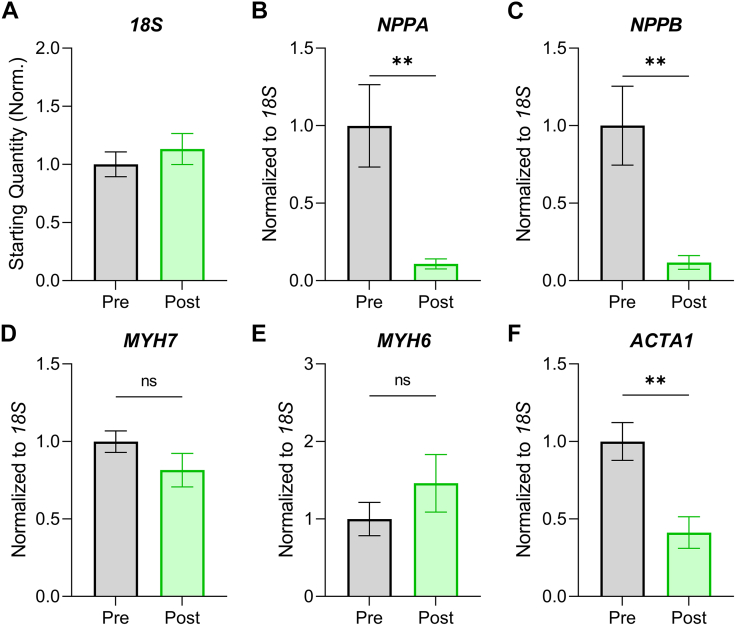
To examine the effect of LVAD placement on pathological gene expression, we used qPCR for common hypertrophy gene markers normalized to 18S ribosomal RNA, the latter of which was unchanged post-LVAD (Figure 2A). We found the expression of NPPA (encoding atrial natriuretic factor) and NPPB (encoding B-type natriuretic peptide) was elevated at the time of initial presentation and was markedly reduced following LVAD (Figures 2B and 2C). Assessment of alpha (MYH6) and beta (MYH7) myosin heavy chain isoform expression suggested a return toward normal isoform ratios (increased MYH6), although the effects were not statistically significant (Figures 2D and 2E). Expression of the skeletal muscle actin isoform (ACTA1) decreased significantly post-LVAD (Figure 2F), further suggesting a restoration of normal gene expression patterns in these patients.
Figure 2.
Pathological Gene Expression Signatures Decrease Following LVAD Therapy
(A) qPCR analysis of 18S ribosomal RNA expression expressed as starting quantity normalized to the pre–left ventricular assist device (LVAD) group. (B) Atrial natriuretic factor (NPPA) gene expression normalized to 18S. (C) B-type natriuretic peptide (NPPB) gene expression normalized to 18S. (D) β-myosin heavy chain (MYH7) gene expression normalized to 18S. (E) α-myosin heavy chain (MYH6) gene expression normalized to 18S. (F) α-skeletal muscle actin (ACTA1) gene expression normalized to 18S. n = 16 pre-LVAD, 16 post-LVAD. Data are presented as mean ± SEM and were analyzed by 2-tailed paired Student's t-test; ∗∗P < 0.01, ns = not significant. Outliers >2 SDs from the mean were removed.
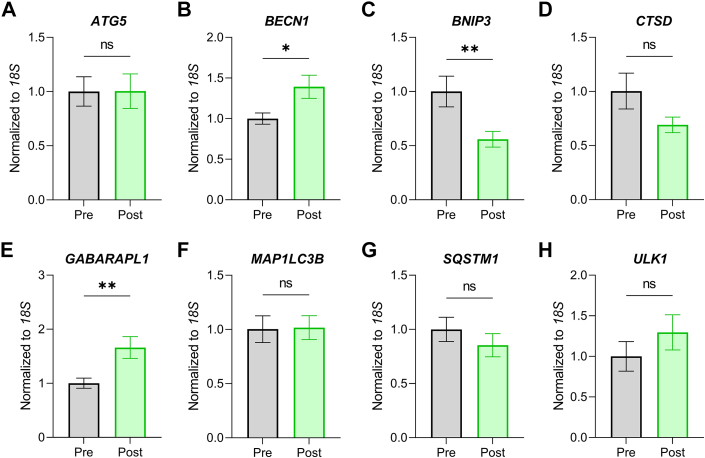
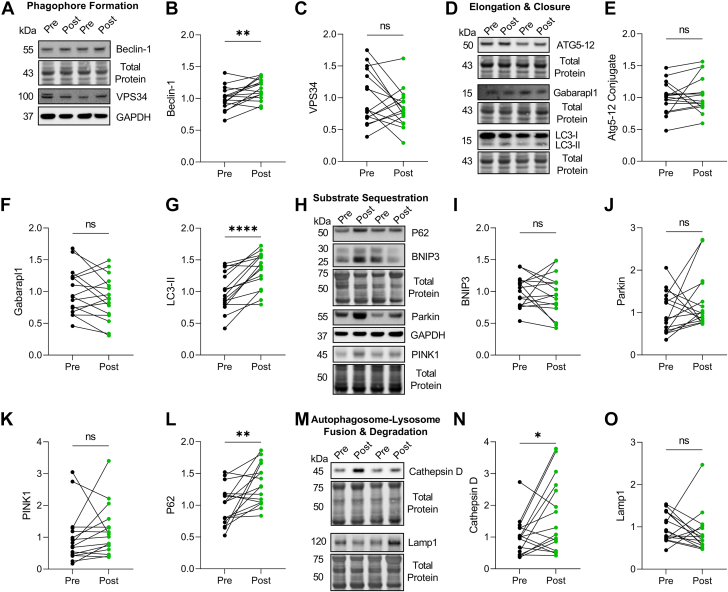
We next assessed the expression of 8 common autophagy genes. No change was found for ATG5, CTSD, MAP1LC3B, SQSTM1, or ULK1 post-LVAD (Figure 3). However, we found that BECN1 and GABARAPL1 gene expression increased, whereas BNIP3 decreased, post-LVAD (Figure 3). To form a more complete picture of autophagy at the protein level, we performed western blots for key autophagy proteins at each stage of the pathway: phagophore formation, elongation/closure, substrate sequestration, and lysosomal degradation. We found increased expression of Beclin-1, LC3-II, P62, and cathepsin D post-LVAD, indicating increased autophagosome number and lysosomal enzyme abundance (Figure 4).
Figure 3.
Autophagy Gene Expression Is Altered Post–Left Ventricular Assist Device
(A)ATG5 gene expression normalized to 18S. (B)BECN1 gene expression normalized to 18S. (C)BNIP3 gene expression normalized to 18S. (D) CTSD gene expression normalized to 18S. (E)GABARAPL1 gene expression normalized to 18S. (F)MAP1LC3B gene expression normalized to 18S. (G)SQSTM1 gene expression normalized to 18S. (H)ULK1 gene expression normalized to 18S. n = 16 pre–left ventricular assist device, 16 post–left ventricular assist device. Data are presented as mean ± SEM and were analyzed by 2-tailed paired Student's t-test; ∗P < 0.05, ∗∗P < 0.01, ns = not significant. Outliers >2 SDs from the mean were removed.
Figure 4.
Autophagy Protein Expression Increases Post–Left Ventricular Assist Device
(A) Representative Western blots for Beclin-1 and VPS34. (B and C). Normalized expression quantification for Beclin-1 (B) and VPS34 (C). (D) Representative Western blots for ATG5-12 conjugate, Gabarapl1, and LC3-II. (E to G) Normalized expression quantification for ATG5-12 (E), Gabarapl1 (F), and LC3-II (G). (H) Representative Western blots for P62, BNIP3, Parkin, and PINK1. (I to L). Normalized expression quantification for P62 (I), BNIP3 (J), Parkin (K), and PINK1 (L). (M). Representative western blots for cathepsin D and Lamp1. (N, O). Normalized expression quantification for cathepsin D (N) and Lamp1 (O). For all, expression was normalized to either GAPDH or Total Protein, n = 16 pre–left ventricular assist device, 16 post–left ventricular assist device; data were analyzed by two-tailed paired Student's t-test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001, ns = not significant
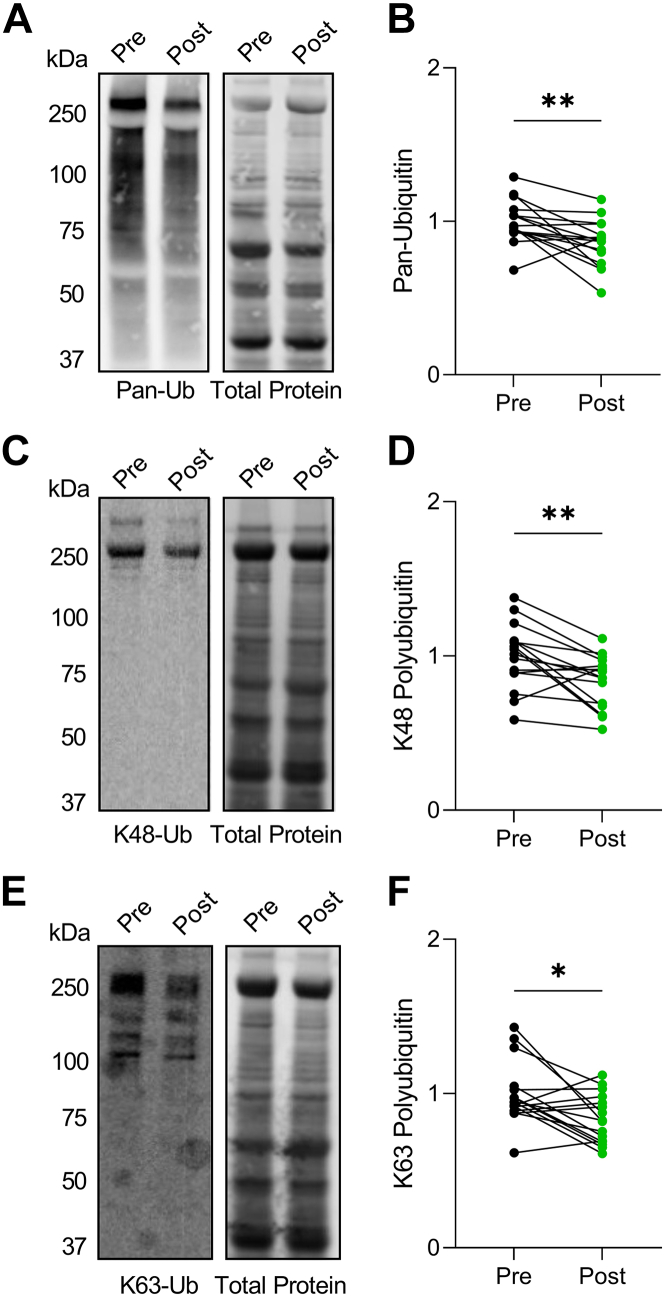
Protein substrates destined for degradation are tagged with a polyubiquitin chain.16 The 2 most common chain linkages in cells are K48 and K63 polyubiquitin. Both linkages may target a protein for autophagic turnover, however, K63 is generally considered to be more specific to autophagy.16 We first probed for total ubiquitin using a pan-ubiquitin antibody and found that ubiquitinated proteins, which increase in dilated cardiomyopathy caused by impaired removal mechanisms,17,18 decreased following LVAD implantation (Figures 5A and 5B). Further exploration into K48 and K63 ubiquitin chains revealed that both decreased post-LVAD (Figures 5C and 5F), supporting that removal of protein substrates destined for degradation by autophagy is enhanced following unloading.
Figure 5.
Ubiquitinated Protein Levels Decrease Post–Left Ventricular Assist Device
(A and B) Representative Western blot for pan-ubiquitin pre- and post-LVAD (A) and quantification normalized to total protein signal (B). (C and D) Representative Western blot for K48-linked poly-ubiquitination pre- and post-LVAD (C) and quantification normalized to total protein signal (D). (E and F) Representative Western blot for K63-linked poly-ubiquitination pre- and post-LVAD (E) and quantification normalized to total protein signal (F). For all, n = 16 pre–left ventricular assist device, 16 post–left ventricular assist device; data analysis by 2-tailed paired Student's t-test; ∗P < 0.05, ∗∗P < 0.01.
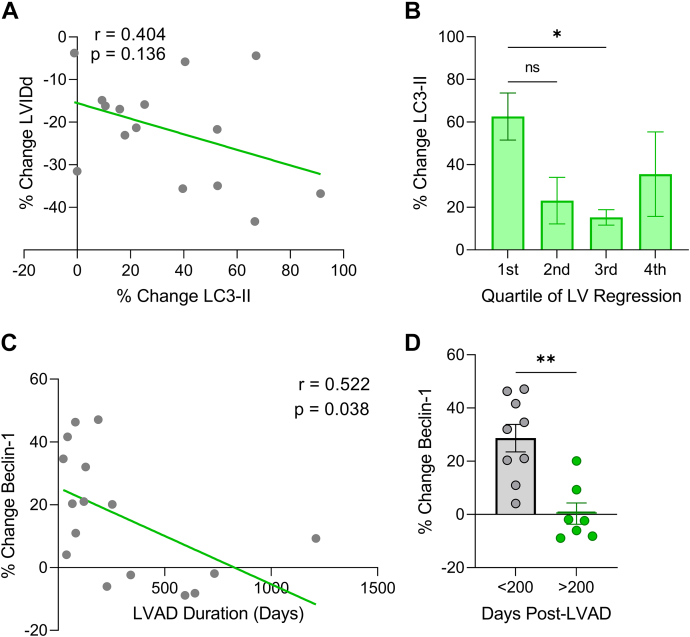
We next examined whether the change in protein expression of key autophagy markers (LC3-II and Beclin-1) correlated with LV structural changes or LVAD treatment duration. We found the LC3-II expression increase post-LVAD was highest in patients who experienced the greatest reduction in LV chamber diameter (Figures 6A and 6B), supporting the possible direct involvement of autophagy in mediating reverse remodeling. Interestingly, the increase in Beclin-1 protein levels post-LVAD declined with duration of LVAD therapy (Figures 6C and 6D), suggesting that increased nascent phagophore synthesis is an early response to unloading that slows with time. Higher levels of autophagy in the acute period following unloading are expected, as the heart must rapidly adjust to new physiological demands before attaining a new equilibrium. None of the other proteins analyzed displayed LVAD duration-dependent expression changes.
Figure 6.
Extent of Increase in Autophagy Proteins LC3-II and Beclin-1 Post-LVAD Is Greatest in Patients With Most Reverse Remodeling and Shortest LVAD Duration, Respectively
(A) % change in LVIDd vs % change LC3-II protein expression post-LVAD analyzed by linear regression. (B) % change LC3-II protein expression post-LVAD organized by quartile of LV regression; first quartile = greatest regression; data are mean ± SEM and were analyzed by 1-way analysis of variance with Tukey’s post hoc test. (C) % change in Beclin-1 protein expression and duration of LVAD therapy analyzed by linear regression. (D) Post-LVAD % change in Beclin-1 expression comparing <200 and >200 days on LVAD; n = 9 <200 days, 7 >200 days; mean ± SEM; 2-tailed Student’s t-test; ∗P < 0.05, ∗∗P < 0.01, ns = not significant. Abbreviations as in Figure 1.
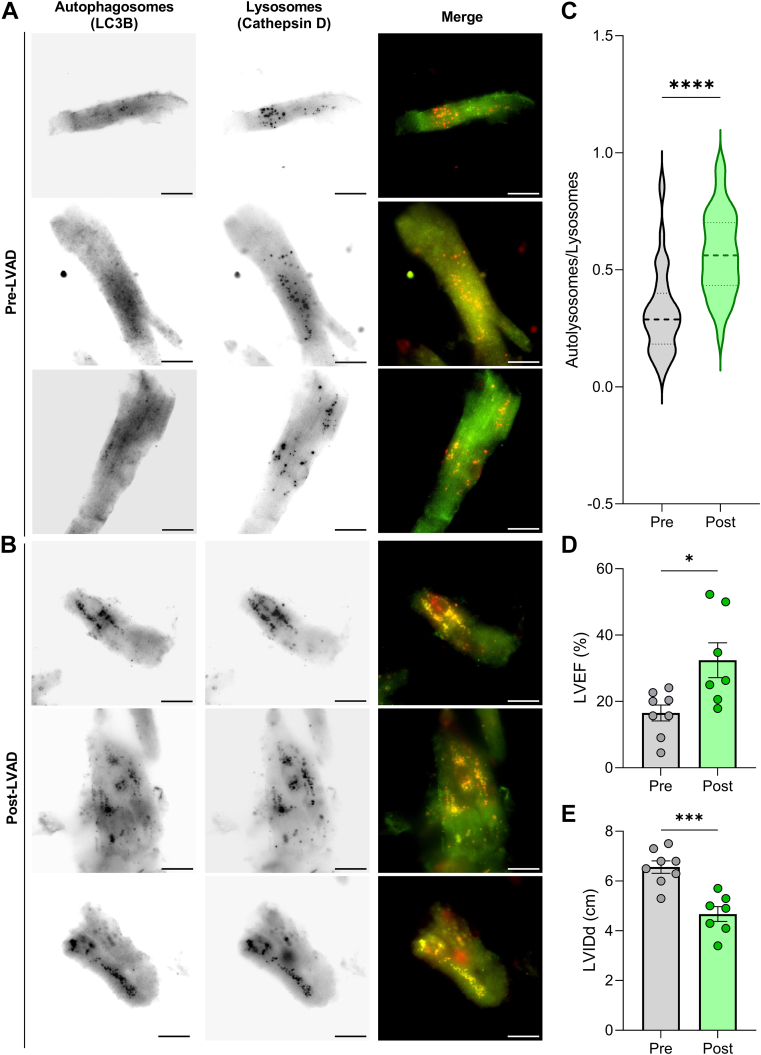
As discussed, increased levels of LC3-II do not necessarily indicate increased autophagy activity. LC3-II levels simply reflect the number of mature autophagosomes, which may increase because of either increased autophagosome synthesis or impaired lysosomal degradation of autophagic substrates. Therefore, to further determine whether autophagy indeed increased post-LVAD, we used immunofluorescence microscopy for LC3 and the lysosome marker cathepsin D in cardiomyocytes from 8 pre- and post-LVAD samples. Colocalization of LC3 with cathepsin D indicates autolysosomes, which represent the active degradation of autophagosome contents (Figures 7A and 7B). Using this method, we identified a significant increase in the proportion of total lysosomes that could be classified as autolysosomes post-LVAD (Figure 7C). Taken together with the increased levels of Beclin-1, LC3-II, and cathepsin D and the decrease of ubiquitinated proteins identified by Western blot, these data strongly support that autophagy increases post-LVAD. These samples were not paired because of limited tissue amounts, but their clinical features were representative of the study population (Figures 7D and 7E).
Figure 7.
Autolysosomes Increase in Post-LVAD Cardiomyocytes
(A and B) Representative immunofluorescence microscopy images of cardiomyocytes from pre-LVAD tissue (A) and post-LVAD tissue (B) stained for LC3B (autophagosomes: green) and cathepsin D (lysosomes: red). 40× magnification, scale bars = 25 μm. (C) Autolysosomes per cell divided by total lysosomes from 8 pre-LVAD samples and 8 post-LVAD samples; n = 93 pre-LVAD, 83 post-LVAD; dark dashed lines denote median values. (D and E) LVEF (D) and LVIDd (E) in the pre- and post-LVAD samples; n = 8 pre, 7 post (data unavailable for 1 patient); mean ± SEM. For all, data were analyzed by 2-tailed Student’s t-test; ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Abbreviations as in Figure 1.
To form a more complete picture of proteolysis post-LVAD, we next assessed the impact of LVAD use on other cellular protein degradation pathways. Using Western blot analysis, we examined the expression of 2 atrophy-associated E3-ubiquitin ligases involved in proteasomal degradation: Atrogin-1 and MuRF-1 (Supplemental Figure 1A). While Atrogin-1 levels were unchanged post-LVAD (Supplemental Figure 1B), MuRF-1 expression displayed a mild but significant increase (Supplemental Figure 1C). We also examined the activity of the 20S proteasome and found that it significantly increased post-LVAD (Supplemental Figure 1D). Enhanced removal of proteasomal clients is further supported by the decreased K48 ubiquitin (Figures 6C and 6D), which is the canonical mechanism of targeting substrates to this proteolytic pathway.19 Activity of the calcium-activated proteases calpain 1 and calpain 2, which partially digest protein components destined for the proteasome,18 also increased post-LVAD (Supplemental Figure 1E).
It is important to consider other factors in patient treatment that might impact the parameters measured. In addition to LVAD therapy, following device implantation, this patient cohort was administered significantly more angiotensin-converting enzyme (ACE) inhibitors (56.3% post-LVAD vs 6.3% pre-LVAD) and beta-blockers (68.8% post vs 12.5% pre). These therapies represent potential contributing factors to the changes observed because each has been independently linked to LV hypertrophy regression in prior studies.3 We therefore analyzed post-LVAD data in groups segregated by coadministered therapies. We found no differences in any parameters (clinical features or protein expression/enzyme activity) for beta-blockers. However, patients who received ACE inhibitors experienced significantly greater improvements in LVEF and LV internal diameter during diastole and greater reduction of ubiquitinated proteins post-LVAD (Supplemental Figure 2). No dose-dependent differences in these parameters were observed; however, this study was not powered to definitively rule out dosage effects (Supplemental Figure 2).
Discussion
We examined autophagy markers in 16 paired pre- and post-LVAD samples. LVAD therapy in these patients was associated with a significant decrease in LV chamber diameter, which positively correlated with improved LVEF. Pathological gene expression signatures also decreased following unloading by LVAD, in agreement with previous findings.20 Examination of autophagy gene/protein expression identified increased Beclin-1 (gene and protein), LC3-II (protein), P62 (protein), and cathepsin D (protein), along with decreased ubiquitinated proteins. Using immunofluorescence microscopy, we further found that autolysosome number increases post-LVAD. Our data indicate widespread changes in autophagy machinery post-LVAD and suggest that activation of this proteolytic pathway increases following therapy (Central Illustration).
Our findings suggesting that autophagy increases post-LVAD are discordant with a previous study on 9 LVAD samples.11 Differences in clinical features between the patient populations may help to explain the apparent incongruencies. Our study included patients who exhibited greater reverse remodeling (25% reduction in LV internal diameter during diastole vs 14%). This cohort also had a higher proportion of Caucasian patients (87.5% vs 56.0%) and longer mean LVAD duration (299 days vs 214 days). Other obvious reasons for the observed difference between the studies are not apparent. In the present study, LC3-II levels increased together with an increase of autolysosomes, suggesting that both autophagosome production and degradation were up-regulated post-LVAD. One unexpected finding was that of increased P62 expression, which is often used to indicate impaired autophagy.21 However, P62 levels have been known to increase in parallel with autophagy activity in some cases.22 Because we see a parallel increase in expression of the lysosomal protease cathepsin D, decrease of ubiquitinated proteins, and increase in autolysosomes, we interpret our collective data to indicate that autophagic activity increases post-LVAD. Of note, increased cathepsin D expression was previously found to be an independent predictor of reverse remodeling in heart disease patients treated with pharmacological therapies.10
The findings presented here on the ubiquitin-proteasome system and calpains agree with and further advance the findings of previously published work. Our data corroborate the findings of 2 previous studies on pre- and post-LVAD tissue where an increase in 20S proteasome activity was identified post-LVAD.11,23 The ubiquitin-proteasome system and E3 ligase MuRF-1 were also implicated in LV mass regression from mechanical unloading by studies in rodent models.24,25 The present study is the first to test activity of the calpain proteases in pre- and post-LVAD samples. An earlier study found calpain gene expression increased post-LVAD but did not assess activity.26 Our findings indicate that calpain activity does in fact increase post-LVAD and agrees with earlier studies on mouse models of mechanical unloading,26,27 suggesting this feature is consistent across species.
One unexpected finding from our study was that improved LVEF after LVAD only occurred in patients who were cotreated with ACE inhibitors. ACE inhibitor use was also associated with significantly better regression of LV chamber diameter. Previous clinical studies have shown that renin-angiotensin-aldosterone system blockade, including with ACE inhibitors, improves reported quality of life, heart failure class, and LV function for patients with LVADs.28 Our data showing that ACE inhibitor use in this patient population is associated with restored proteostasis (ie, reduction of ubiquitinated protein levels) may provide mechanistic insight into the underlying biology. It is therefore important to note that the high prevalence of ACE inhibitors used on our patient cohort post-LVAD may be a contributing factor to the molecular features observed. This might also explain some differences observed between this and the previous study, where the proportion of patients on ACE inhibitors was not reported.11
Study limitations
Human tissue studies come with many technical challenges. One applicable to the present study is the inability to monitor autophagy markers over time in a single patient, which would inform how this process changes directly after unloading compared with months/years later. However, our data with Beclin-1 suggests that autophagy may be most up-regulated in the acute period following unloading. Another shortcoming is that true autophagic flux cannot be measured in tissue biopsies, because this would require injection of an autophagy inhibitor (eg, Bafilomycin) before tissue collection, which would be unethical. Future studies using preclinical heart failure models to decipher the acute and long-term autophagic flux response to mechanical unloading will be highly valuable. Ultrastructural analyses of autophagy would also be beneficial. Unfortunately, these were not possible in the present study because of limited tissue quantities from the pre-LVAD samples. Another limitation is that, despite having 16 paired samples, this study was underpowered to fully examine potential sex differences because only 4 of the patients were female. Large clinical studies have identified sex differences in reverse remodeling from first-line therapies, including LVAD, where female patients typically display more favorable responses.3 Future studies focused on the molecular changes between men and women are therefore warranted. The subgroup analyses based on ACE inhibitor administration are also limited and would greatly benefit from larger sample sizes to examine the dose-dependence of these effects. Furthermore, because of low sample size, we are unable to perform any analysis based on when medications were discontinued.
Conclusions
Clinical studies spanning nearly 40 years indicate that many pharmacological, surgical, and device therapies can induce LV reverse remodeling in a subset of patients.3 Importantly, this reverse remodeling is often associated with partial myocardial recovery and improved patient quality of life.3 However, because of a lack of molecular-level studies in human samples, the cellular features underlying this favorable process are poorly understood. LVAD therapy is associated with the most robust reverse remodeling of any currently administered treatment.3 Our findings indicate that reverse remodeling after LVAD is associated with increased autophagy. This aligns with the recent finding that autophagy is up-regulated in patients who display reverse remodeling when treated with common pharmacological therapies.10 Together with a wealth of literature in preclinical heart failure models showing that autophagy-activating compounds effectively prevent adverse remodeling and induce its regression,29, 30, 31, 32, 33 these studies in human samples suggest that targeting autophagy may be of therapeutic value for patients with heart disease.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Reverse remodeling of the LV in heart disease patients is closely linked to improved clinical and quality of life outcomes. LVAD therapy commonly induces reverse remodeling in patients with end-stage heart failure, and this morphological change is associated with partial to near-complete myocardial recovery in many patients. The relationship between regression of LV hypertrophy and improved outcomes has led to the idea that directly targeting adverse remodeling with compounds that induce its reversal may be efficacious. However, before this becomes reality, a comprehensive understanding of the molecular pathways that drive this process is required. In this study, we analyzed protein degradation pathways (primarily autophagy) to examine how these are impacted in end-stage nonischemic heart failure patients who received LVADs as a bridge-to-transplant. Our findings support the conclusion that cellular protein degradation pathways, including autophagy, have increased activity post-LVAD. Previous studies in rodents identified a direct role for autophagy in hypertrophy regression, and our findings suggest that the same may be true in human hearts.
TRANSLATIONAL OUTLOOK: The molecular factors that mediate cardiac tissue mass regression remain poorly defined. Our findings support that mechanical unloading by LVAD therapy in human hearts activates the autophagy pathway, in agreement with recently published work from rodent models of mechanical LV unloading. First, LVAD therapy was associated with increased expression of the autophagy proteins Beclin-1 and LC3-II, canonical markers of autophagy activation. Second, the increased abundance of autophagosomes coincided with increased expression of the lysosomal protease cathepsin D and increased number of autolysosomes—denoting the active degradation of autophagosome contents by lysosomes. Finally, the levels of K63 polyubiquitinated proteins (autophagy substrates) were reduced following LVAD, indicating that proteins destined for autophagy that accumulate in heart disease are readily removed with post-LVAD autophagy activation. These findings add support for the importance of autophagy in the heart and its dysregulation in disease. Moreover, these findings highlight the need for future investigations on the therapeutic potential of targeting autophagy in heart disease.
Funding Support and Author Disclosures
The authors have received support from the National Institute of Health (R01HL117138 and R01GM029090 to Dr Leinwand, T32 HL007822 to Dr Martin, and T32 GM142607 to Dr Juarros). REDCap was provided by National Institutes of Health/NCATS Colorado CTSA Grant Number UL1 TR002535. The contents are the authors’ sole responsibility and do not necessarily represent the official views of the National Institutes of Health. Dr Leinwand is a cofounder of MyoKardia, acquired by Bristol Myers Squibb; and is a paid member of their Scientific Advisory Board. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
Daniel Burkhoff, MD, served as Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures and a table, please see the online version of this paper.
Appendix
References
- 1.Goldstein D.J., Naka Y., Horstmanshof D., et al. Association of clinical outcomes with left ventricular assist device use by bridge to transplant or destination therapy intent: the Multicenter Study of MagLev Technology in Patients Undergoing Mechanical Circulatory Support Therapy with HeartMate 3 (MOME) JAMA Cardiol. 2020;5:411–419. doi: 10.1001/jamacardio.2019.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkhoff D., Topkara V.K., Sayer G., Uriel N. Reverse remodeling with left ventricular assist devices. Circ Res. 2021;128:1594–1612. doi: 10.1161/CIRCRESAHA.121.318160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin T.G., Juarros M.A., Leinwand L.A. Regression of cardiac hypertrophy in health and disease: mechanisms and therapeutic potential. Nat Rev Cardiol. 2023;20(5):347–363. doi: 10.1038/s41569-022-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 5.Shirakabe A., Zhai P., Ikeda Y., et al. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133:1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Q., Qu C., Niu T., et al. Nrf2-mediated cardiac maladaptive remodeling and dysfunction in a setting of autophagy insufficiency. Hypertension. 2016;67:107–117. doi: 10.1161/HYPERTENSIONAHA.115.06062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hariharan N., Ikeda Y., Hong C., et al. Autophagy plays an essential role in mediating regression of hypertrophy during unloading of the heart. PLoS One. 2013;8 doi: 10.1371/journal.pone.0051632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans S., Ma X., Wang X., et al. Targeting the autophagy-lysosome pathway in a pathophysiologically relevant murine model of reversible heart failure. J Am Coll Cardiol Basic Trans Science. 2022;7:1214–1228. doi: 10.1016/j.jacbts.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyabu J., Yamaguchi O., Hikoso S., et al. Autophagy-mediated degradation is necessary for regression of cardiac hypertrophy during ventricular unloading. Biochem Biophys Res Commun. 2013;441:787–792. doi: 10.1016/j.bbrc.2013.10.135. [DOI] [PubMed] [Google Scholar]
- 10.Kanamori H., Yoshida A., Naruse G., et al. Impact of autophagy on prognosis of patients with dilated cardiomyopathy. J Am Coll Cardiol. 2022;79:789–801. doi: 10.1016/j.jacc.2021.11.059. [DOI] [PubMed] [Google Scholar]
- 11.Kassiotis C., Bailai K., Wellnitz K., et al. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation. 2009;120:S191–S197. doi: 10.1161/CIRCULATIONAHA.108.842252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishida K., Otsu K. Autophagy during cardiac remodeling. J Mol Cell Cardiol. 2016;95:11–18. doi: 10.1016/j.yjmcc.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Schiattarella G.G., Hill J.A. Therapeutic targeting of autophagy in cardiovascular disease. J Mol Cell Cardiol. 2016;95:86–93. doi: 10.1016/j.yjmcc.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Cui T. Autophagy modulation: a potential therapeutic approach in cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2017;313:H304–H319. doi: 10.1152/ajpheart.00145.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin T.G., Tawfik S., Moravec C.S., Pak T.R., Kirk J.A. BAG3 expression and sarcomere localization in the human heart are linked to HSF-1 and are differentially affected by sex and disease. Am J Physiol Heart Circ Physiol. 2021;320(6):H2339–H2350. doi: 10.1152/ajpheart.00419.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z., Popelka H., Lei Y., Yang Y., Klionsky D.J. The roles of ubiquitin in mediating autophagy. Cells. 2020;9(9):2025. doi: 10.3390/cells9092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin T.G., Myers V.D., Dubey P., et al. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun. 2021;12(1):2942. doi: 10.1038/s41467-021-23272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin T.G., Kirk J.A. Under construction: The dynamic assembly, maintenance, and degradation of the cardiac sarcomere. J Mol Cell Cardiol. 2020;148:89–102. doi: 10.1016/j.yjmcc.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grice G.L., Nathan J.A. The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci. 2016;73:3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaxall B.C., Tschannen-Moran B.M., Milano C.A., Koch W.J. Differential gene expression and genomic patient stratification following Left Ventricular Assist Device support. J Am Coll Cardiol. 2003;41:1096–1106. doi: 10.1016/s0735-1097(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 21.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18(9):1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q., Su H., Ranek M.J., Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wohlschlaeger J., Sixt S.U., Stoeppler T., et al. Ventricular unloading is associated with increased 20s proteasome protein expression in the myocardium. J Hear Lung Transplant. 2010;29:125–132. doi: 10.1016/j.healun.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 24.Cao D.J., Jiang N., Blagg A., et al. Mechanical unloading activates FoxO3 to trigger Bnip3-dependent cardiomyocyte atrophy. J Am Heart Assoc. 2013;2(2) doi: 10.1161/JAHA.113.000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis M.S., Rojas M., Li L., et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296:H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razeghi P., Volpini K.C., Wang M.E., Youker K.A., Stepkowski S., Taegtmeyer H. Mechanical unloading of the heart activates the calpain system. J Mol Cell Cardiol. 2007;42:449–452. doi: 10.1016/j.yjmcc.2006.08.114. [DOI] [PubMed] [Google Scholar]
- 27.Liang L., Li H., Cao T., et al. Calpain activation mediates microgravity-induced myocardial abnormalities in mice via p38 and ERK1/2 MAPK pathways. J Biol Chem. 2020;295:16840–16851. doi: 10.1074/jbc.RA119.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedrzejewska A., Braczko A., Kawecka A., et al. Novel targets for a combination of mechanical unloading with pharmacotherapy in advanced heart failure. Int J Mol Sci. 2022;23(17):9886. doi: 10.3390/ijms23179886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juric D., Wojciechowski P., Das D.K., Netticadan T. Prevention of concentric hypertrophy and diastolic impairment in aortic-banded rats treated with resveratrol. Am J Physiol Heart Circ Physiol. 2007;292:H2138–H2143. doi: 10.1152/ajpheart.00852.2006. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg T., Abdellatif M., Schroeder S., et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–1438. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Chen C., Yao F., et al. AMPK inhibits cardiac hypertrophy by promoting autophagy via mTORC1. Arch Biochem Biophys. 2014;558:79–86. doi: 10.1016/j.abb.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Sciarretta S., Forte M., Castoldi F., et al. Caloric restriction mimetics for the treatment of cardiovascular diseases. Cardiovasc Res. 2021;117:1434–1449. doi: 10.1093/cvr/cvaa297. [DOI] [PubMed] [Google Scholar]
- 33.Sciarretta S., Yee D., Nagarajan N., et al. Trehalose-induced activation of autophagy improves cardiac remodeling after myocardial infarction. J Am Coll Cardiol. 2018;71:1999–2010. doi: 10.1016/j.jacc.2018.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.