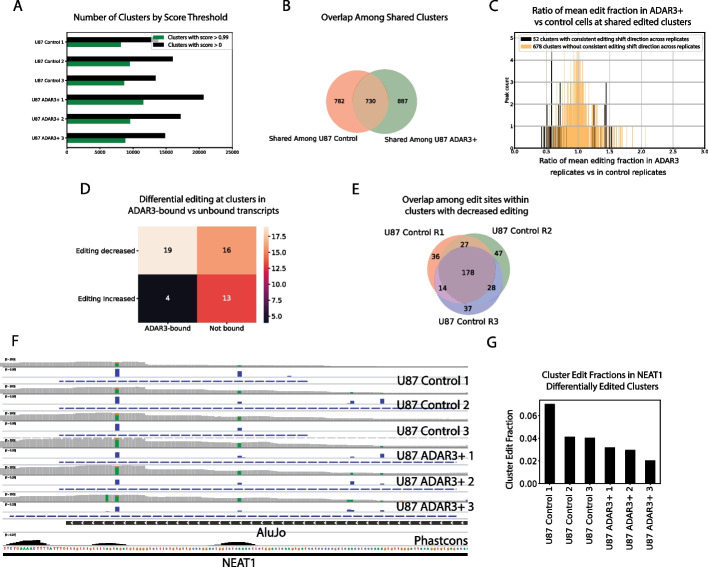
Fig. 4.
FLARE identifies regions with ADAR3-linked differential A-to-I editing. a Number of A-to-I editing clusters called across samples, without and with minimum score filtering. b Shared replicated clusters distribution. c Histogram depicting ratio of mean edit fraction in ADAR3+ vs control U87 at shared editing clusters identified by FLARE. d Shared clusters bound by ADAR3 are highly enriched for a decrease in edit fraction in the ADAR3+ condition compared to the control condition. e Even among clusters shared by replicates, many editing sites are unique to each replicate. f IGV snapshot of a Alu-overlapping region towards the 3’ end of non-coding RNA NEAT1 which experiences reduced editing in an ADAR3+ context. Coverage, edit fraction, and cluster regions plots are shown for each sample, with edits highlighted on bar plots (“A”s are green and “G”s are brown). A phastcons (20 species) conservation track is also shown at bottom. g Edit fractions across samples, within the differentially edited NEAT1 cluster