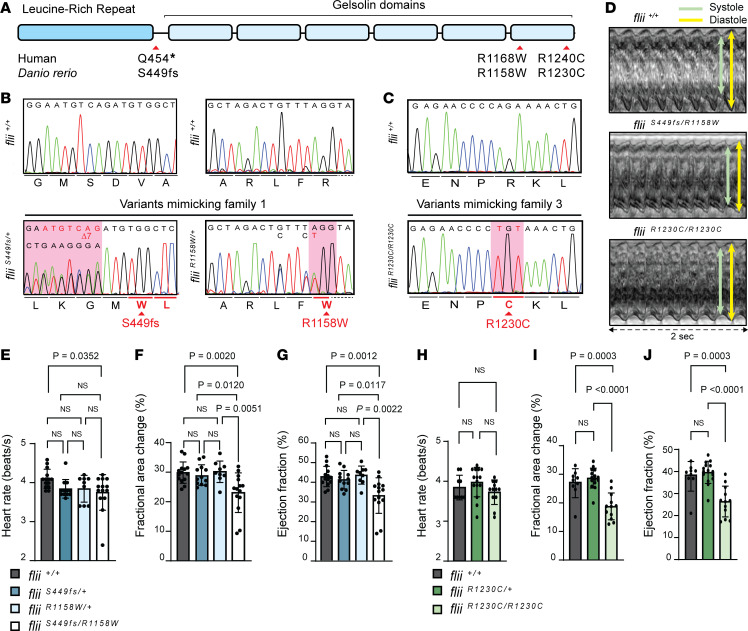
Figure 2. CRISPR/Cas9-mediated genome editing of patient-specific biallelic variants in flii in zebrafish results in DCM-associated phenotypes in early development.
(A) Schematic representation of the FLII protein, consisting of a leucine-rich repeat (LRR) and 6 gelsolin-like domains. Location of the human variants of families 1 and 3 are depicted with corresponding variants generated in zebrafish (D. rerio). (B) Aligned Sanger sequencing traces of wild-type flii+/+ and genome-edited PCR amplicons from compound heterozygous fliiS449fs/R1158W larvae harboring a heterozygous variant resulting in a frameshift starting at amino acid position 449 (left panel, sequence is reverse complement as it was sequenced with the reverse primer) and the heterozygous R1158W missense variant (right panel), representing family 1. Modified codons are underlined in red. Dotted line represents intronic sequence. (C) Representative Sanger sequencing results of the PCR amplicons from wild-type and fliiR1230C/R1230C zebrafish larvae harboring the homozygous R1230C missense variant, representing family 3. The modified codon is underlined in red. (D) Ventricular kymographs derived from high-speed imaging video recordings of 120 hpf zebrafish larvae spanning approximately 2 seconds: flii+/+ (top panel), fliiS449fs/R1158W (middle panel) and fliiR1230C/R1230C (lower panel). Note that there are no signs of irregular heart rhythm in larvae harboring patient-specific biallelic variants. (E–G) Ventricular contractility parameters derived from high-speed imaging movies, including heart rate (E), fractional area change (F), and ejection fraction (G) for flii+/+, fliiS449fs/+, fliiR1158W/+, and fliiS449fs/R1158W. flii+/+ n = 14; fliiS449fs/+ n = 12; fliiR1158W/+ n = 9; fliiS449fs/R1158W n = 14. (H–J) Ventricular contractility parameters derived from high-speed imaging movies, including heart rate (H), fractional area change (I), and ejection fraction (J) for flii+/+, fliiR1230C/+, and fliiR1230C/R1230C. flii+/+ n = 9; fliiR1230C/+ n = 15; fliiR1230C/R1230C n = 12. Statistics: mean ± SD; 1-way ANOVA coupled with Tukey’s multiple-comparison test was used to test for significance.