Abstract
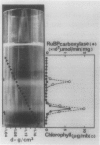
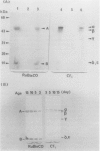
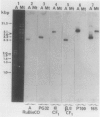
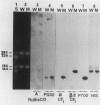
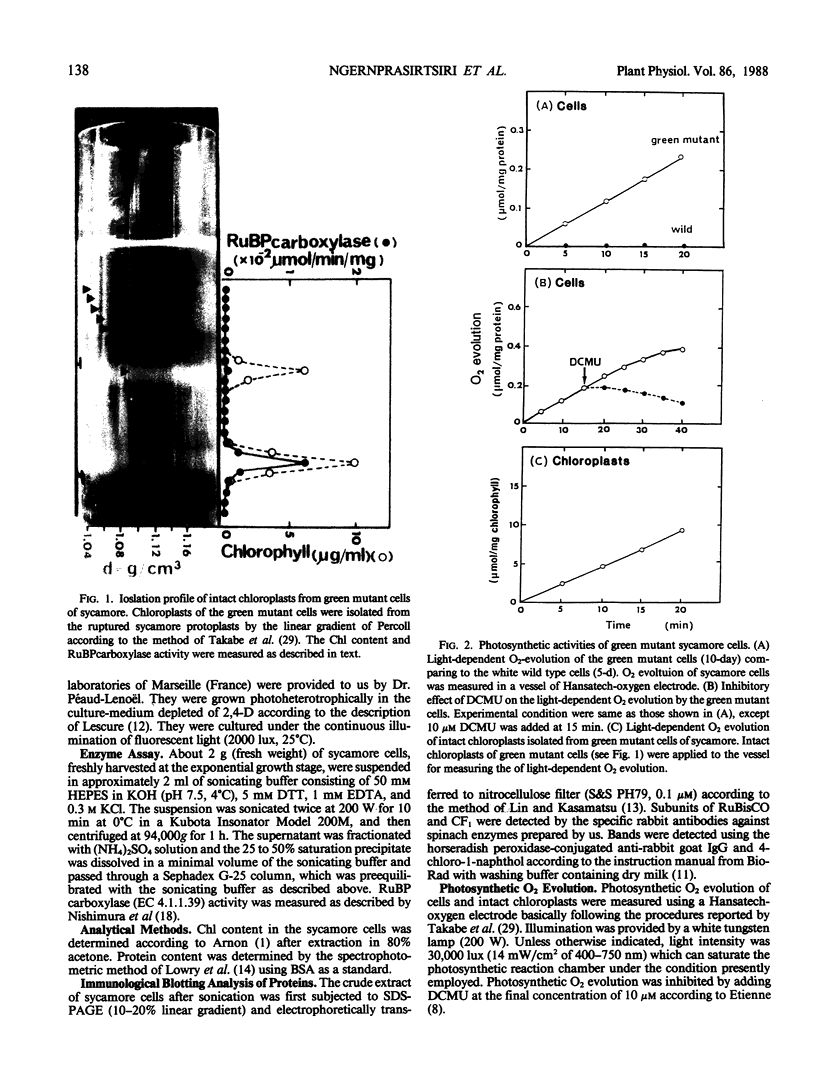
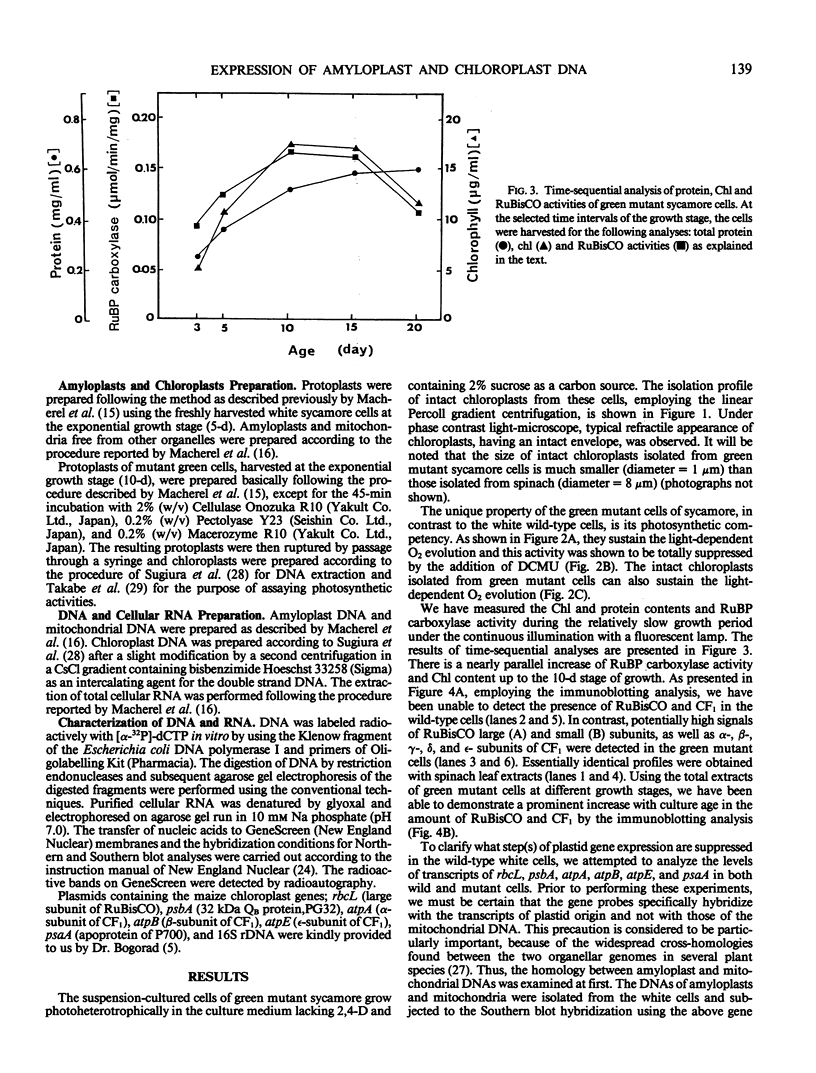
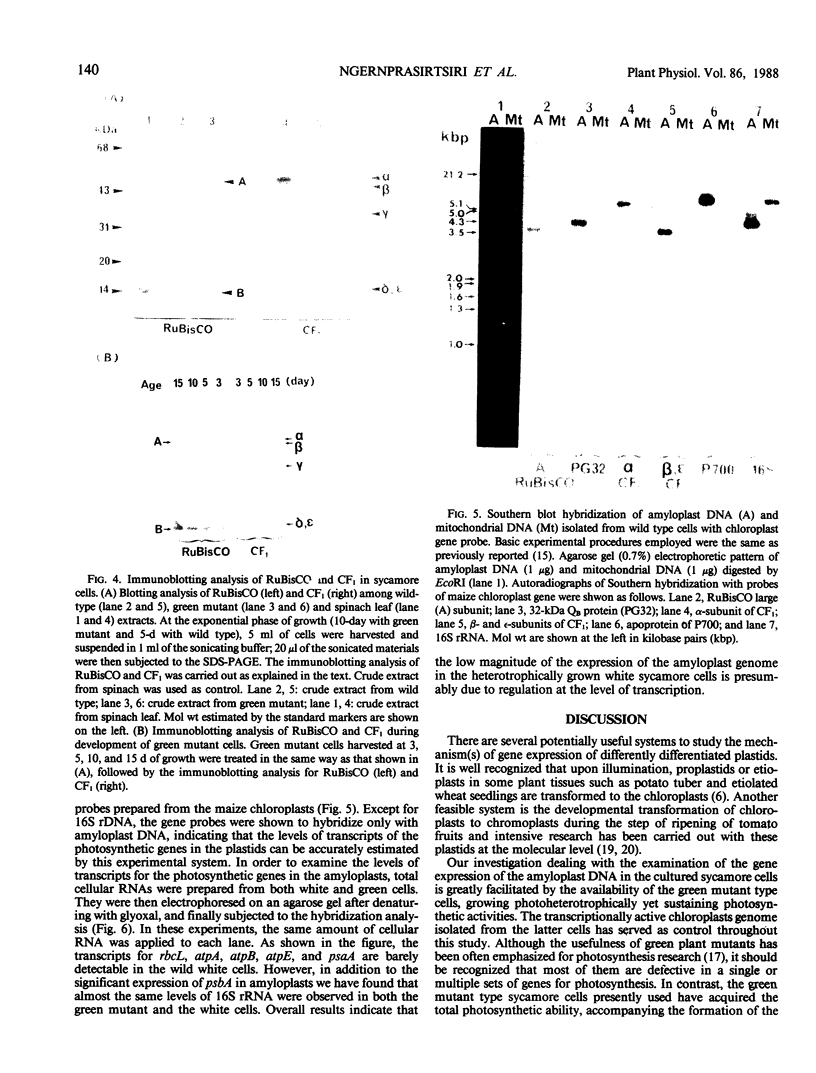
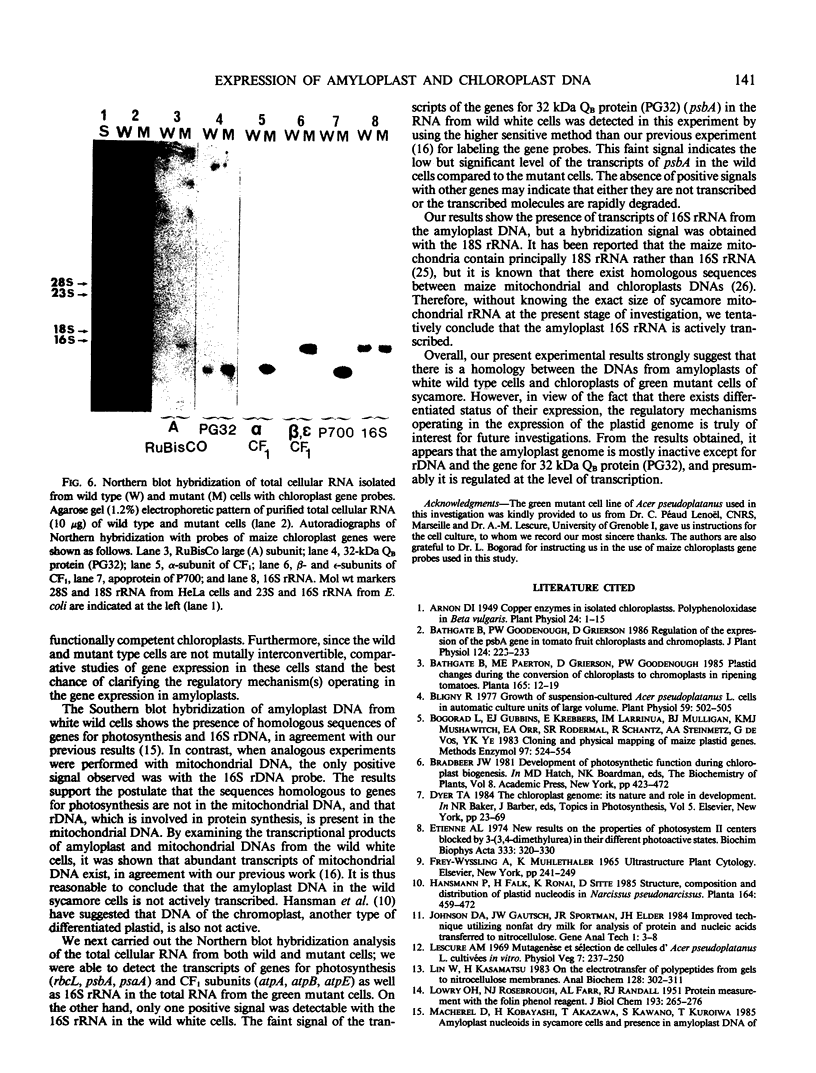
Green mutant cells of sycamore (Acer pseudoplatanus L.), which had been selected by mutagenic treatment of the white wild type, grow photoheterotrophically in auxin-depleted culture medium. In contrast to the wild-type cells, mutant cells exhibit photosynthetic O2-evolution activity during their growth coincident with increases of (a) chlorophyll, (b) protein, and (c) ribulose-1,5-bisphosphate (RuBP) carboxylase activity. Functionally competent chloroplasts were isolated from the green cells. Mechanism(s) governing gene expression of amyloplast DNA in the heterotrophically grown white cells were compared with those of the chloroplast DNA isolated from the mutant cells. We have demonstrated in both amyloplast and chloroplast DNAs the presence of sequences homologous to the maize chloroplast genes for photosynthesis, including the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO)(rbcL), the 32 kDa QB protein (PG32) (psbA), the apoprotein of P700 (psaA) and subunits of CF1 (atpA, atpB, and atpE). However, employing either enzyme assays or immunological techniques, RuBisCO and CF1 cannot be detected in the white wild type cells. Northern blot hybridization of the RNA from the white cells showed high levels of transcripts for the 16S rRNA gene and low level of transcripts for psbA; based on comparison with results obtained using the green mutant cells, we propose that the amyloplast genome is mostly inactive except for the 16S rRNA gene and psbA which is presumably regulated at the transcriptional level.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnon D. I. COPPER ENZYMES IN ISOLATED CHLOROPLASTS. POLYPHENOLOXIDASE IN BETA VULGARIS. Plant Physiol. 1949 Jan;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligny R. Growth of Suspension-cultured Acer pseudoplatanus L. Cells in Automatic Culture Units of Large Volume. Plant Physiol. 1977 Mar;59(3):502–505. doi: 10.1104/pp.59.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lin W., Kasamatsu H. On the electrotransfer of polypeptides from gels to nitrocellulose membranes. Anal Biochem. 1983 Feb 1;128(2):302–311. doi: 10.1016/0003-2697(83)90379-2. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Dyer T. A., Lonsdale D. M. Organization of the mitochondrial ribosomal RNA genes of maize. Nucleic Acids Res. 1982 Jun 11;10(11):3333–3340. doi: 10.1093/nar/10.11.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Lonsdale D. M. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature. 1982 Oct 21;299(5885):698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]