Abstract
Background
Histologically assessed hepatocyte ballooning is a key feature discriminating nonalcoholic steatohepatitis (NASH) from steatosis (NAFL). Reliable identification underpins patient inclusion in clinical trials and serves as a key regulatory-approved surrogate endpoint for drug efficacy. High inter/intra-observer variation in ballooning measured using the NASH-CRN semi-quantitative score has been reported yet no actionable solutions have been proposed.
Methods
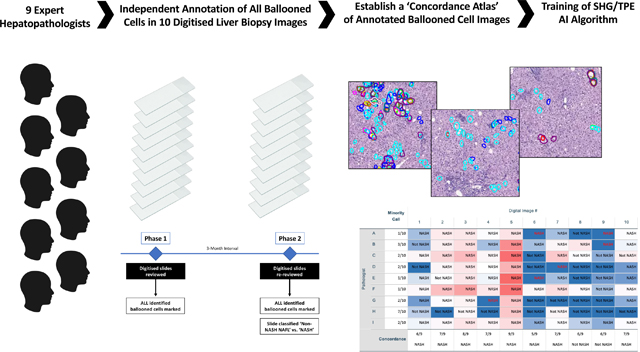
A focussed evaluation of hepatocyte ballooning recognition was conducted. Digitised slides were evaluated by 9 internationally recognized expert liver pathologists on two separate occasions: each pathologist independently marked every ballooned hepatocyte and later provided an overall non-NASH NAFL/NASH assessment. Interobserver variation was assessed and a ‘concordance atlas’ of ballooned hepatocytes generated to train second harmonic generation/two-photon excitation fluorescence imaging-based artificial intelligence (AI).
Results
Fleiss kappa statistic for overall interobserver agreement for presence/absence of ballooning was 0.197 (95%CI 0.094–0.300), rising to 0.362 (0.258–0.465) with a ≥5-cell threshold. However, intraclass correlation coefficient for consistency was higher (0.718 [0.511–0.900]), indicating ‘moderate’ agreement on ballooning burden. 133 ballooned cells were identified using a ≥5/9 majority to train AI ballooning detection (AI-pathologist pairwise concordance 19–42%, comparable to inter-pathologist pairwise concordance of between 8–75%). AI quantified change in ballooned cell burden in response to therapy in a separate slide set.
Conclusions
The substantial divergence in hepatocyte ballooning identified amongst expert hepato-pathologists suggests that ballooning is a spectrum, too subjective for its presence or complete absence to be unequivocally determined as a trial endpoint. A concordance atlas may be used to train AI assistive technologies to reproducibly quantify ballooned hepatocytes that standardise assessment of therapeutic efficacy. This atlas serves as a reference-standard for ongoing work to refine how ballooning is classified by both pathologists and AI.
Keywords: Nonalcoholic fatty liver disease, Nonalcoholic steatohepatitis, NASH, NAFLD, Ballooning, Artificial intelligence, Machine learning, Histology
LAY SUMMARY
For the first time, we show that, even amongst expert hepatopathologists, there is poor agreement about number of ballooned hepatocytes seen in the same digitized histology images. This has important implications as presence of ballooning is needed to establish the diagnosis of nonalcoholic steatohepatitis (NASH), and its unequivocal absence is one of the key requirements to show ‘NASH resolution’ to support drug efficacy in clinical trials. Artificial intelligence-based approaches may provide a more reliable way to assess the range of injury recorded as “hepatocyte ballooning” as a clinical trial endpoint.
Graphical abstract
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) covers a pathological spectrum of liver injury characterized by excess fat accumulation within hepatocytes in the absence of harmful alcohol consumption [1, 2]. NAFLD encompasses steatosis (nonalcoholic fatty liver, NAFL), steatohepatitis (nonalcoholic steatohepatitis, NASH), fibrosis and ultimately cirrhosis [3]. Being highly prevalent, it places a substantial burden on global healthcare resources that is predicted to increase further over the next decade [4, 5]. Consequently, there is substantial interest and a need to develop pharmacological therapy.
Although grade (activity) of steatohepatitis waxes and wanes over time [6, 7], it is accepted as the underlying driver of fibrogenesis [6], which in turn determines long-term outcome [7, 8]. Therefore, current FDA and EMA regulatory guidance mandates that drug development should target patients with NASH rather than NAFL, as the latter may be best addressed through lifestyle change [9, 10]. This distinction is key to patient selection for trial enrolment and also serves as one of the surrogate endpoints for drug efficacy assessment [9, 10]. Histological assessment of liver biopsy remains the basis for diagnosing NASH, grading activity and assessing stage of fibrosis. The presence of hepatocellular ballooning is generally considered an essential component in the composite of histological features leading to a diagnosis of NASH as it is thought to represent a form of hepatocyte injury associated with fibrogenesis that is not seen in non-progressive disease [11]. Two semiquantitative scoring systems have been proposed to aid consistent histopathological interpretation and grading and staging of biopsies: the NASH-Clinical Research Network (CRN) ‘NAFLD Activity Score’ (NAS) and fibrosis stage; and the FLIP/EPoS ‘Steatosis-Activity-Fibrosis’ (SAF) score [12, 13]. Both measure hepatocyte ballooning on a 3-point scale (0–2) but with nuanced differences. It is apparent, however, that the categorical definitions in both semi-quantitative systems may be subject to variation in their interpretation and application. No study to date has specifically addressed ballooning changes at the individual cell level through evaluation and annotation of high-resolution digitized images.
Interobserver variation in pathologists’ assessment of grade of activity in general, and ballooning specifically, are documented. Kappa values of 0.56–0.57 for application of NAS ballooning score in two separate studies based on light microscopic analyses from the Pathology Committee of the NASH CRN have been published almost 15-years apart [12, 14]. Another, more recent, interobserver study also highlighted the discordance of assessment of all features of NASH, including ballooning (linearly weighted kappa for ballooning 0.517) [15]. The implication of this being that trial entry criteria had only been met in 53.7% of biopsies re-read at the end of the study [15]. These reported levels of inter- and intra-observer agreement are a cause for concern. Since regulators place great emphasis on ballooning as a requisite feature of NASH in clinical trials, it may affect study recruitment and assessment of drug efficacy, with potentially deleterious consequences for drug development pipelines and impeding patient access to efficacious treatments.
There is a pressing need for reproducible, objective and standardized evaluation of the significant histopathological features that discriminate NAFL from NASH, in particular, presence and quantification of hepatocyte ballooning. Recognition of this need is evidenced by the development and move towards early adoption of artificial intelligence (AI) algorithms to support histopathological assessment, particularly from digitized slide review [16–18]. Development of these tools necessitates a detailed understanding of what features define hepatocyte ballooning and how these are perceived, interpreted and applied in practice by expert hepato-pathologists.
The primary goals of the current study were: firstly, to utilize input from blinded independent assessments by nine internationally recognized expert hepatopathologists to generate a dataset of reliably and reproducibly identified ballooned hepatocytes that can be used to support the development of machine learning (artificial intelligence) algorithms for the detection and quantification of hepatocyte ballooning; and secondly, to conduct a focused study that accurately evaluated interobserver variation in hepatocyte ballooning feature recognition. Digitized slides were chosen because they are increasingly used in clinical trials and because only digitisation facilitates the necessary granular annotation of individual cells.
MATERIAL & METHODS
Composition of the expert-pathology group
Nine internationally recognized expert hepatopathologists from the USA (EMB, ZG, CG, DEK, MY), Europe (CL, DGT), Australia (ADC) and Singapore (AW) participated. All were senior pathologists with extensive experience in assessing NAFLD and applying the NASH-CRN NAS scoring system in routine practice and in the clinical trial setting.
Histology Samples
This study utilized liver biopsy samples from two randomized controlled trials (Seladelpar trial from CymaBay Therapeutics, Inc [NCT03551522]), and Resmetirom trial from Madrigal Pharmaceuticals, Inc. [NCT02912260]) [19, 20]. The ‘development’ cohort comprised ten trial-entry screening biopsies selected to encompass a spectrum of NAFLD grade/stage from non-NASH NAFL (i.e. no ballooning, B0) to NASH with marked ballooning (B2) and moderate fibrosis (F2–3). Twenty-two cases with paired biopsies were selected from the Resmetirom Phase 2 NASH trial as an independent ‘test’ cohort for the qBallooning2 algorithm. Digitised images of the H&E stained liver tissue sections were acquired using the Aperio Digital Pathology Imaging Systems (Leica Biosystems). Detailed descriptions of the samples and processes are provided in Supplementary Methods & Data.
Process for Biopsy Evaluation
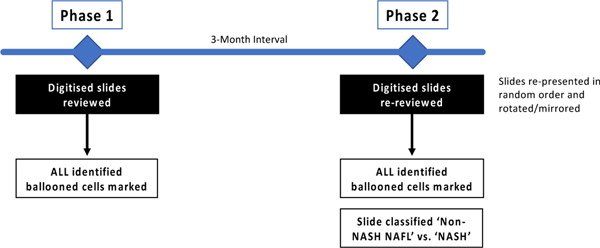
After an initial period to gain familiarity with the web-based histology platform by examining and marking a large number of practice slides over an 8-week period, data acquisition for the study was conducted in two phases temporally separated by a 3-month interval (Figure 1). Pathologists performed the tasks independently and without knowledge of the group’s results until completion of the study. Selection of the regions of interest that were used in this study was done by a single expert hepatopathologist in order to: (1) normalize the area of liver tissue to be analysed as the biopsies varied in length and number of cores; (2) encompass a range of ballooning from none to many, as in “real life” in practice and in clinical trials; and (3) cover a range of technical biopsy preparation (ie staining) quality, also as in “real life”.
Figure 1.
In Phase 1, ten digital pathology images were reviewed by individual pathologists, circling all ballooned cells. In Phase 2, after an interval of 3-months, the same images rotated through 90 degrees/mirrored were re-presented in a different order. Additionally, pathologists were asked to report for each slide if they considered it diagnostic of NASH vs. non-NASH NAFL.
Phase 1:
10 pre-selected regions of interest were extracted from the digital slides, as described above, for scoring ballooning. Pathologists were instructed to circle all ballooned hepatocytes within the digital biopsy slide images and were aware that the annotation would be used to enable the assessment of interobserver agreement for ballooned cell identification. For fields that contained overlapping ballooned hepatocytes, the pathologists were instructed to circle the entire cluster if they were not be able to define individual cells using their best efforts.
Phase 2:
After an interval of 3 months, the same 10 slides were re-presented to the pathologists in a different random order and with some of the images rotated through 90 degrees or mirrored. Pathologists were not informed that these were the same images previously assessed or that rotation or mirroring had occurred. Pathologists were asked to report for each slide if they considered it diagnostic of NASH vs. non-NASH NAFL. Additionally, to allow intra-observer variation to be assessed, pathologists were also instructed to circle all ballooned hepatocytes on three of the images using the same criteria as they had applied during Phase 1.
SHG/TPEF Microscopy & qBallooning2 algorithm development
All imaging of unstained sections was conducted by trained technicians on identical equipment (Genesis™ system HistoIndex Pte. Ltd., Singapore) according to a standardized operating procedure. Detailed descriptions of the protocols are provided in Supplementary Methods & Data.
Annotated ballooned cells on the 10 pre-selected digital H&E slides made by the pathologists during Phase 1 were recorded and used to generate the “ground truth” of training sets on the corresponding SHG/TPEF images for the artificial intelligence algorithm. Suitable candidates of ballooned hepatocytes on the TPEF channel were identified using traditional image analysis methods, including image segmentation, morphological processing, and watershed algorithm as previously described [16].
A total of 45 ballooning parameters were established and quantified, including the number of ballooned hepatocytes, the area of ballooned hepatocytes and the area of “collagen area” around the ballooned hepatocytes. Subsequently, paired digitized liver biopsy slides (n = 44) from the development set were used to establish a qBallooning2 index, which can indicate the degree of ballooning. Images were processed and analysed using MATLAB 8.3 (The MathWork, USA).
Statistics Analysis
The number of annotated cells per slide as annotated by each pathologist were both quantified and data collected on cells annotated by more than one pathologist. Data were collated in Microsoft Excel and analysis performed using SPSS v.26 (IBM Inc. USA). Considering number of ballooned hepatocytes as a continuous variable, the single-measures intraclass correlation coefficient (ICC) for absolute agreement and consistency was tested [21]. Inter/intraobserver agreement was then assessed for three binary target conditions: (i) Presence of any hepatocyte ballooning; (ii) Presence of at least 5 ballooned hepatocytes; and (iii) ‘non-NASH NAFL’ vs. ‘NASH’ using Fleiss’ kappa statistic [22]. Ballooned hepatocyte counts were also transformed to generate a three-point semi-quantitative ballooning score (SQBS) (0–2) to align with both NAS and SAF methods according to the number of ballooned hepatocytes per image reported by each pathologist. SQBS was defined as 0= <5; 1= 5–75; 2 = >75, with the cutoff between SQBS 1 and 2 derived from the overall mean + 1SD of the number of ballooned cells reported per slide. The consistency of SQBS among pathologists was calculated using pairwise linear weighted kappa statistics. The thresholds for kappa interpretation proposed by Landis and Koch were applied [23]. Difference of changes for qBallooning2 continuous values was calculated by Wilcoxon rank sum test. Statistical significance level was set at p < 0.05 throughout.
Data Availability
The raw and annotated images used in this study are presented in the Supplementary Image File.
RESULTS
Based on counting nuclei, the mean (±SD) number of hepatocytes examined and classified as ballooned, or, by default, not ballooned per slide by each pathologist was 8,150 (±3,378) for each of the 10 biopsies studied (Supplementary Table S1). Histological images demonstrating ballooned hepatocyte mark-up for all slides examined at Phase 1 are provided in Supplementary Image File. A significant difference in the mean number of ballooned hepatocytes identified per slide was observed (ANOVA F(9,80) = 16.69, p<0.0005) supporting the successful a priori selection of cases to represent a range of ballooned cell burden.
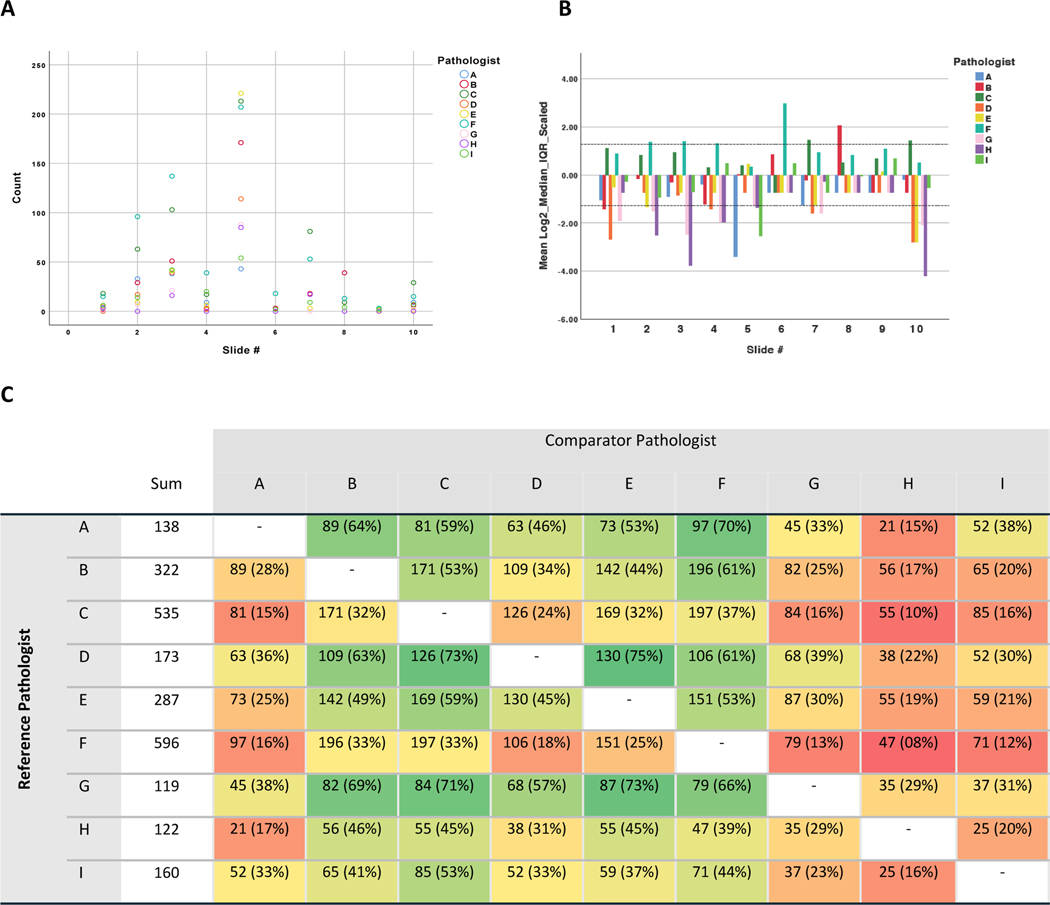
At Phase 1, it was apparent that there was substantial interobserver variation in the number of hepatocytes identified as being ballooned across the majority of the images studied (Figure 2A and Supplementary Tables S1 and S2). Image #5 was considered to demonstrate the greatest degree of ballooning, although it also showed the greatest range in number of ballooned cells reported (mean 133 cells, range 43–221). This remained true when pathologist concordance was considered (Supplementary Table S3). Image #9 had the least and the narrowest range of readings (mean 1, range 0–3). When at Phase 2 a sub-set of the images were rotated and blindly re-evaluated to identify all visible ballooned hepatocytes, on average 54.6% of cells identified by a given pathologist at Phase 1 were again identified as ballooned by the same pathologist at Phase 2 (range 32% to 91%), Supplementary Table S4.
Figure 2.
A: Count of Cells Circled on Each Image by Pathologist. It is notable that in all but two images (#3 and #5), at least one pathologist reported no ballooned cells present. Zero ballooning was agreed on by two pathologists in two images (#4 and #7); five pathologists in two images (#8 and #9), and six pathologists in one (#6). All pathologists except two (F and I) recorded zero ballooning at least once. B: Scaled Count of Cells Circled by Slide and Pathologist demonstrating pathologist propensity to identify hepatocytes as ballooned. Pathologist F systematically reported greater numbers of ballooned cells than the majority of their peers, followed by C. In contrast, pathologists G, H, and D systematically reported less ballooning. C: Heatmap showing Pairwise Agreement in Cells Identified as Ballooned between Pathologists. Pairwise agreement in ‘ballooned cell’ call, n (%), where percentage refers to the proportion of cells identified as ballooned by the reference pathologist that were also identified by the comparator pathologist. Heatmap shaded to denote percentage interobserver agreement relative to the reference pathologist (green = high, red = low).
Overall Interobserver Agreement on Ballooned Hepatocytes
As detailed in the methods section, interobserver agreement amongst pathologists on hepatocyte ballooning was assessed for three target conditions: (i) Presence of any hepatocyte ballooning; (ii) Presence of at least 5 ballooned hepatocytes; and (iii) Concordance of a Semi-Quantitative Ballooning Score (SQBS).
The overall level of interobserver agreement amongst pathologists for the presence of any hepatocyte ballooning was classed as ‘poor’ with a Fleiss kappa statistic of 0.197 (95%CI 0.094–0.300, p<0.0005). If a threshold of detecting at least 5 ballooned cells was applied to consider ballooning present, this rose to attain a ‘fair’ level of agreement (kappa 0.362, 95%CI 0.258–0.465, p<0.0005).
Considering number of ballooned hepatocytes as a continuous variable, the single-measures intraclass correlation coefficient (ICC) for absolute agreement was of a similar level at 0.640 (95%CI 0.410–0.864, p<0.0005) indicating ‘low-moderate’ levels of interobserver agreement. Whilst ICC consistency levels were slightly higher (0.718, 95%CI 0.511–0.900, p<0.0005), indicating that there was ‘moderate’ agreement on those cases that exhibited broadly greater or lesser numbers of ballooned cells, the levels of concordance for identifying the same specific cells as ballooned in pairwise comparison between pathologists varied substantially (range between 8% to 75%, Figure 2C).
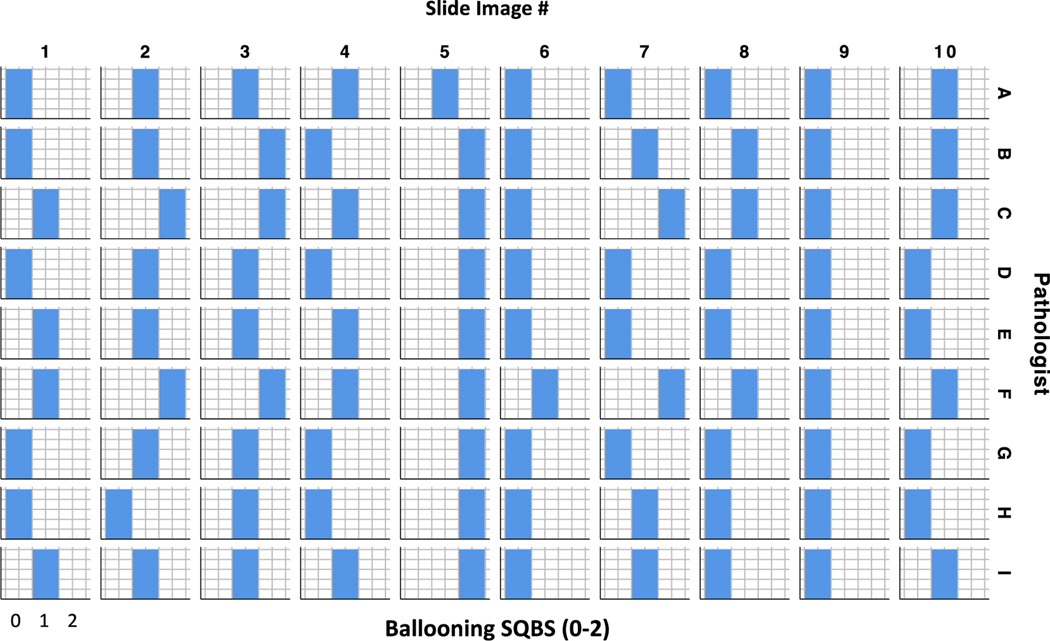
In light of this we modelled the performance of a semiquantitative scoring system derived from absolute number of ballooned hepatocytes using arbitrary thresholds, ballooning hepatocyte counts were transformed to generate a three-point semi-quantitative ballooning score (SQBS) (0–2) to align with both NAS and SAF methods according to the number of ballooned hepatocytes per image reported by each pathologist. SQBS was defined as 0= < 5; 1= 5–75; and 2 = > 75 ballooned hepatocytes reported per slide. Figure 3 summarises the SQBS score for each slide image by pathologist. Comparing SQBS categories, interobserver pairwise weighted kappa values ranged between 0.231 – 1.000 (Supplementary Table S5), suggesting some pathologists were more closely aligned in their broad quantification of hepatocyte ballooning than others. However, overall, the level of interobserver agreement between pathologists remained only ‘fair’ (kappa 0.291, 95%CI 0.210–0.371, p<0.0005). Although there was substantial variation at the cell level between Phase 1 and Phase 2, levels of intraobserver agreement based on SQBS for three digital images were broadly similar to interobserver agreement, kappa values ranging between 0.250–1.000 with 5 pathologists achieving intraobserver kappa values of 1.000 between the two phases.
Figure 3.
SQBS Ballooning derived from absolute Ballooned Cell count per slide for each pathologist. The calculated SQBS category is shown by individual pathologist for each slide image (SQBS Ballooning 0 <5 cells circled; 1= 5–75; 2 = > 75).
Cell-level interobserver agreement
In light of the variation in the absolute number of ballooned cells reported per slide (Figure 2C), and the apparent divergence as to which individual cells pathologists deemed to be ballooned on each image, we sought to identify patterns of interpretation amongst pathologists and factors that influence determination of hepatocyte ballooning.
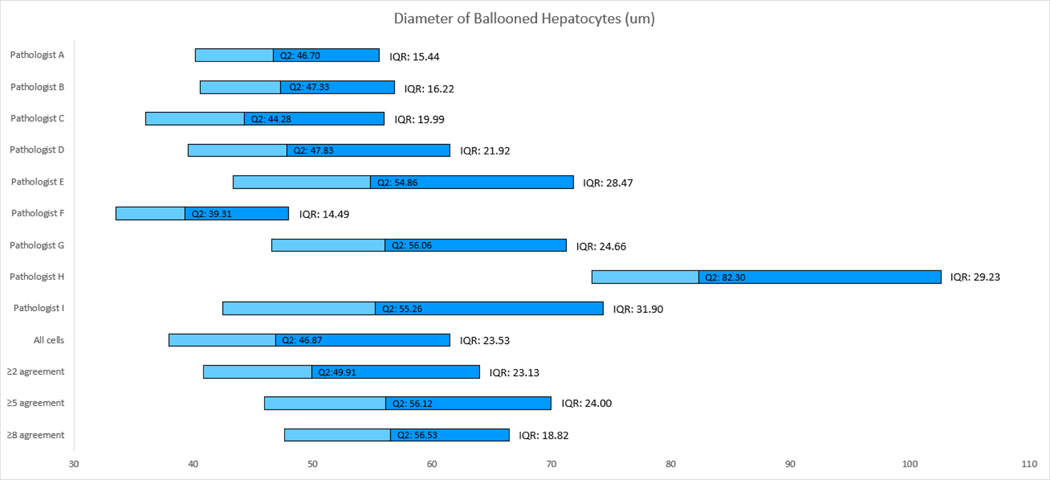
In the absence of a ‘gold standard’ test for ballooning that could provide a ground truth, we hypothesised that the median number of ballooned hepatocytes identified across the expert pathologist group for each image would approximate to the ‘true’ number of ballooned hepatocytes. Sustained deviation from this value across multiple images was used to identify pathologists that tended to report greater or lesser numbers of ballooned hepatocytes than their peers (Figure 2B). Pathologist F systematically reported greater numbers of ballooned hepatocytes than the majority of their peers, followed by C. In contrast, pathologists H, G and D consistently reported fewer cells as ballooned (Figure 2B). To assess how strongly cell size influenced each pathologist’s assessment, the median diameter and interquartile range of the encircled ballooned hepatocytes was calculated for each pathologist (Figure 4). Although significant overlap was observed, it was notable that the pathologists consistently reporting the greatest and least number of cells as ballooned appeared to diverge in how much emphasis they placed on cell size, with those that considered more cells to be ballooned adopting a more permissive, lower, cell-diameter threshold (pathologists F 39.31 ± 14.49μm and C 33.28 ± 19.99μm) compared to those that identified the least cells to be ballooned (pathologist H 82.30 ± 29.23μm), Wilcoxon rank sum test p<0.001. Restricting analysis to larger cells (greater than 2× or 3× normal hepatocytes) however had little effect on interobserver agreement, confirming that adopting a size threshold would not be sufficient to improve interobserver agreement (data not shown).
Figure 4.
Chart based on the lower quartile, median and upper quartile of the nine pathologists and their agreements after removing large clusters. The median and IQR of all ballooned hepatocytes identified by each pathologist.
Relevance of Ballooned Hepatocyte Presence to the Determination of ‘non-NASH NAFL’ vs. ‘NASH’
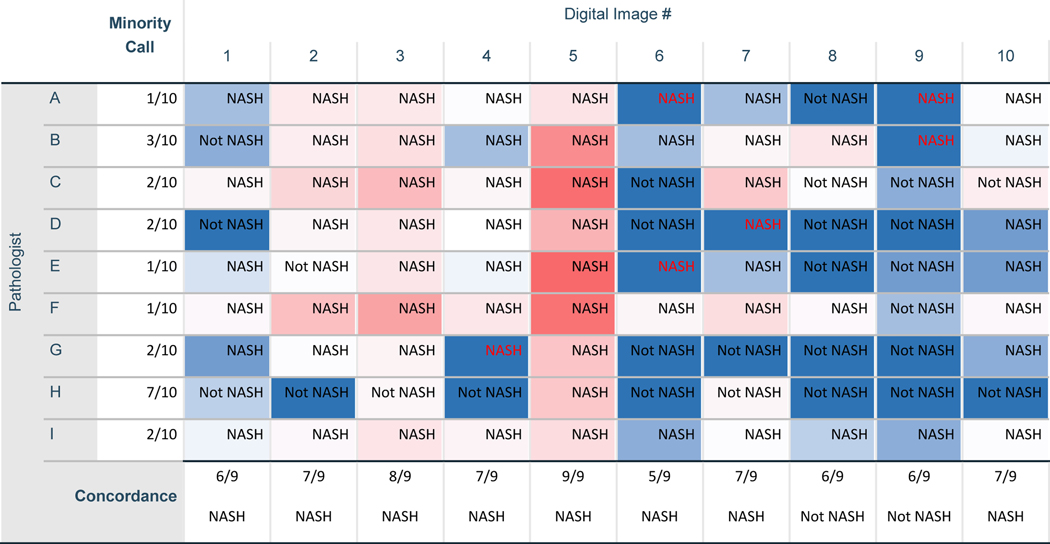
A key requirement for clinical trial recruitment, and as a trial endpoint, is the histological determination of the presence or absence of NASH, i.e. the distinction of ‘non-NASH NAFL’ from ‘NASH’. In the second phase of the study, 3-months after the initial quantification of ballooned hepatocytes, and without access to their previous ballooned cell counting results, the pathologists were asked to re-review each slide image and provide an overall ‘gestalt’ diagnosis of either NASH or non-NASH NAFL based on all histological features observed. Surprisingly, the kappa value for agreement of a NASH diagnosis was just 0.127 (95%CI 0.024–0.230, P=0.016), indicative of ‘little or no agreement’ between the pathologists on the presence or absence of NASH when operating independently. As shown in Figure 5, there was only one image (#5) for which all pathologists agreed that NASH was present, that being the same image in which all pathologists had identified ballooned hepatocytes and 8 of 9 pathologists had previously identified high levels of ballooning (SQBS 2). There were no cases for which all pathologists agreed that NASH was absent. Notwithstanding these high levels of interobserver variation, a majority concordance diagnosis could be ascertained for most images, and at least 7 of the 9 pathologists independently agreed on disease category for six of the ten images (Figure 5). Minority calls were reasonably evenly spread across the pathologists. Although pathologist H did provide a minority opinion in 7/10 cases, excluding this pathologist had a modest effect on the overall kappa value of the group (0.201, 95%CI 0.084–0.318, p<0.001).
Figure 5.
Table cells are coloured Blue through to Red as a heat map indicating the relative number of Ballooned hepatocytes identified by each pathologist (dark blue denotes cases for which a given pathologist has indicated that no ballooned hepatocytes were present at Phase 1. Colour changes through light blue to white and then red as the number of ballooned cells identified increases, with darker red indicating that many ballooned cells were seen). The non-NASH NAFL vs NASH diagnosis at Phase 2 made independently by each pathologist is shown, along with the degree of concordance for this decision (as a fraction out of nine pathologists) and the majority decision for each digital image. Where NASH is shown in red text, this denotes a NASH diagnosis call by a pathologist at Phase 2 despite previously reporting that no Ballooned hepatocytes were present in the digital image during Phase 1.
For only two pathologists was there a significant positive correlation between their determination of ballooning presence and the diagnosis of NASH (pathologists D and G, Phi 0.816, p=0.010 for each). For trial endpoints based on NASH resolution, this implies that there was little correlation between a determination of the absence of ballooning at either the absolute or the <5 cell threshold and the pathologists diagnosing non-NASH NAFL, suggesting that pathologists may also rely on additional features to aid the differential diagnosis between NAFL and NASH. Adopting the majority diagnostic-category opinion for each case as the reference standard, little correlation was observed between the mean number of ballooned hepatocytes reported and whether a diagnosis of NASH was made (Kendall’s tau 0.447, p=0.117). Indeed, there were 6 cases classified as NASH by a given pathologist in which the same pathologist had previously identified zero ballooned cells (Figure 5).
Leveraging a Histological ‘Ground Truth’ Atlas of Hepatocyte Ballooning to develop “qBallooning2”, a novel SHG/TPEF-based machine learning algorithm
Despite the apparent interobserver variation in the identification of ballooned hepatocytes described above, a substantial number of hepatocytes were consistently identified as ballooned, or non-ballooned, by multiple pathologists (Table 1). These constitute a histological ‘ground truth’ annotated cell image atlas in which the rigor of ballooned cell determination may be calibrated according to the degree of concordance (i.e. number of agreeing pathologists) at the individual cell level.
Table 1:
Use of the Histological ‘Ground Truth’ Atlas to tune the qBallooning2 Algorithm
| qBallooning2 training-set cell-selection criteria | Number of ballooned cells Identified by Pathologists | Number of ballooned cells Identified by qBallooning2 | Overlap between qBallooning2 and majority concordance of ≥5-Pathologists | Positive Predictive Value Proportion of ballooned cells called by qBallooning2 are ‘True Positive’ * | False Discovery Rate Proportion of ballooned cells called by qBallooning2 are ‘False Positive’ * | True Positive Rate (Sensitivity) Proportion of ballooned cells identified by qBallooning2 * | False Negative Rate Proportion of ballooned cells missed by qBallooning2* |
|---|---|---|---|---|---|---|---|
| Agreement of any 1 pathologist | 1188 | 346 | 54 | 54/346 (16%) | 292/346 (84%) | 54/133 (41%) | 79/133 (59%) |
| Agreement of at least 2 pathologists | 481 | 250 | 51 | 51/250 (20%) | 199/250 (79.6%) | 51/133 (38%) | 82/133 (62%) |
| Agreement of at least 3 pathologists | 284 | 170 | 37 | 37/170 (22%) | 133/170 (78.2%) | 37/133 (28%) | 96/133 (72%) |
| Agreement of at least 4 pathologists | 188 | 114 | 25 | 25/114 (22%) | 89/114 (78%) | 25/133 (19%) | 108/133 (81%) |
| Agreement of at least 5 pathologists | 133 | 88 | 22 | 22/88 (25%) | 66/88 (75%) | 22/133 (17%) | 111/133 (83%) |
| Agreement of at least 6 pathologists | 86 | 59 | 16 | 16/59 (27%) | 43/59 (73%) | 16/133 (12%) | 117/133 (88%) |
| Agreement of at least 7 pathologists | 59 | 40 | 15 | 15/40 (38%) | 25/40 (62.5%) | 15/133 (11%) | 118/133 (89%) |
| Agreement of at least 8 pathologists | 26 | 24 | 5 | 5/24 (21%) | 19/24 (79%) | 5/133 (4%) | 128/133 (96%) |
Table comparing the performance of qBallooning2 in the development dataset. The algorithm was optimized to detect ballooned cells using data derived from each level of interobserver concordance and shows how the level of interobserver concordance stipulated affects the performance of the algorithm.
Relative to majority concordance of ≥5-pathologists.
By coupling these annotated image data to the associated SHG/TPEF scanned images in the development cohort, we next built upon our previous work to further develop and refine a SHG/TPEF-based machine learning algorithm for ballooned hepatocyte identification [16]. From an overall data set of 45 features (Supplementary Table S6), the enhanced “qBallooning2” index was established based on 7 parameters, including 6 ballooned cell parameters: total perimeter of ballooned hepatocytes per unit tissue area, variance in distance between ballooned hepatocytes and the nearest ballooned hepatocytes, average distance between ballooned hepatocytes and the nearest ballooned hepatocytes, average number of ballooned hepatocytes within 100 μm of a ballooned hepatocyte, variance in number of ballooned hepatocytes within 100 μm of a ballooned hepatocyte; and 1 collagen parameter: total collagen area around ballooned hepatocytes per unit tissue area.
Example images showing how ballooned hepatocytes identified by the expert histopathologists align with those identified by qBallooning2 are shown in Supplementary Figure S1. When qBallooning2 was trained using the full atlas of 1,188 cells identified as ballooned by at least one pathologist, 346 cells were flagged by the algorithm of which 198 cells (57%) had also been identified by the pathologists. Performance of the qBallooning2 algorithm could be further tuned according to the number of pathologists providing concordance that were used to train it. We systematically used all possible training-sets (agreement of ≥1 pathologist, ≥2 pathologists, all the way to ≥8 pathologists) and measured its performance by counting the number of overlapping cells between the algorithm and pathologists’ annotations (Table 1). qBallooning2 had pairwise overlap with individual pathologists ranging from 19% (with Pathologist F) to 42% (with Pathologist G), which was comparable to the level of inter-observer variation between pathologists of 8–75%. Algorithms trained with greater interobserver concordance identified fewer cells and exhibited less sensitivity but tended to better control false discovery rate. This potentially allows the algorithm to be tuned to be more or less conservative according to how it is to be used.
To define a reference standard for comparisons of performance, a concordance threshold of ≥5 pathologists (ie a simple majority) was adopted. Considering first the individual pathologists, this demonstrated sensitivity (true positive rate) ranging between 44–94%, with positive predictive values (PPV) of 13–53% and false discovery rates (FDR) 47–87%, and an estimated specificity (true negative rate) >99%. In comparison, qBallooning2 algorithms trained with atlases containing at least 50 cells exhibited sensitivity ranging between 11–41% (PPV 16–38%, FDR 62–84%) according to how the algorithm was trained, again with specificity >99% (Table 1). It should be noted that specificity is based upon an estimated mean 8,150 cells per slide and will tend to appear high as ballooned cells are an infrequent feature in any biopsy.
The qBallooning2 algorithm that had been optimised using concordance of ≥5-pathologists was selected as an exemplar for further study. The consequent algorithm exhibited a sensitivity of 17% (PPV 25%, FDR 75%).
Demonstration of qBallooning2 quantification in NASH clinical trials
To establish proof-of-principal whether qBallooning2 was sensitive to change in the context of NASH clinical trials, samples obtained from the Resmetirom Phase 2 trial formed an independent test cohort. Samples were chosen from patients that, irrespective of treatment arm, at the end of the study were reported by the trial pathologist to have either at least 1-point NASH-CRN ballooning score reduction (‘improvers’), or no ballooning score reduction (‘non-improvers’).
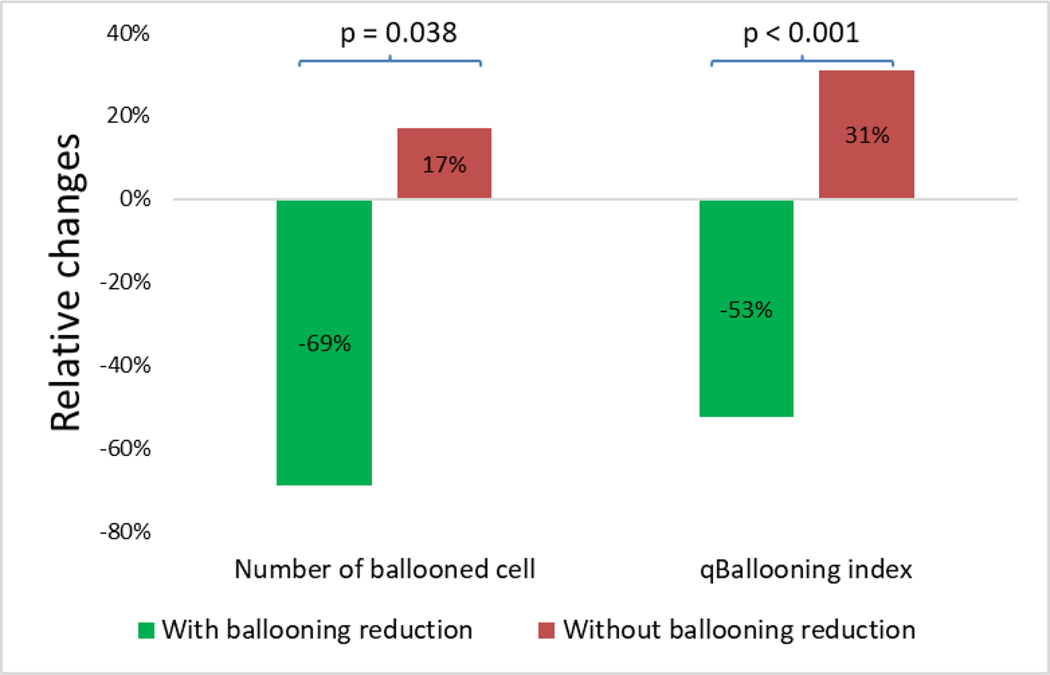
Amongst ‘improvers’ that were judged to show a reduction in ballooned hepatocytes by the trial pathologist, relative to the baseline biopsy qBallooning2 detected a median (lower quartile, upper quartile) 79% (−89%, −19%) reduction in number of ballooned hepatocytes. In contrast, a mean 77% (−46%, 143%) increase in ballooned hepatocytes was detected in ‘non-improvers’ at the end of the study (p=0.038). This was shown with corresponding qBallooning2 indices of −59% (−71%, 20%) and +5% (−25%, 25%) respectively (p=0.008) (Figure 6).
Figure 6.
Note that the ballooning scores used are those that had been issued by the central pathologist of the trial.
DISCUSSION
Histological assessment of liver biopsy has been widely adopted as the reference standard against which performance of therapeutics are assessed. However, there is a growing literature demonstrating considerable inter- and intra-observer variation in the scoring of liver biopsies [12–15, 24]. The presence of hepatocyte ballooning is generally considered a pathognomonic feature that is necessary for a diagnosis of NASH as it is thought to represent a form of liver cell injury associated with fibrogenesis [11]. Although variability in the morphological interpretation of the ballooning feature is recognized (as discussed below) a significant correlation of ballooning with fibrosis progression and prognosis has been described [25]. The ability to accurately diagnose NASH, and by extension also identify its absence in order to fulfil the FDA mandated endpoint of NASH resolution without worsening of fibrosis, hinges on the ability to accurately demonstrate an absence of hepatocyte ballooning [26]. It is therefore of great relevance for drug development both in terms of clinical trial enrolment and also as an efficacy endpoint [6, 27].
A key finding in this study is that, despite many years of cumulative experience, there remains substantial divergence amongst expert hepatopathologists as to which specific cells constitute ballooned hepatocytes (Figure 2C). This was apparent in our study irrespective of whether pathologists had previously spent time collaborating, for example within the NIDDK NASH CRN histopathology group, or not. Whilst the distilled concept of ‘hepatocellular ballooning’ may be appealing, and significantly enlarged ballooned hepatocytes with obvious Mallory-Denk bodies may be more readily identified, in practice pathologists must recognise and interpret multiple visual cues when assessing presence of ballooning. This is supported by the numerous descriptors commonly utilised in the literature [28–31] and may explain why although agreeing verbally and in nearly all written documents [12], in practice there is substantial divergence in how this visual information is assimilated and applied [15]. This was clearly demonstrated in the current analysis in which certain pathologists consistently identified greater or fewer numbers of ballooned hepatocytes and placed greater or lesser emphasis on cell size (Figure 2B and Figure 4). Indeed, the magnitude of the observed variation was sufficient to alter classification when a 3-point semi-quantitative score was applied (Figure 3). These findings suggest that the patterns recognised by pathologists when identifying “hepatocellular ballooning” are based on a variable constellation of hepatocyte features, which may include cell size, cell shape and ill-defined nuclear and cytoplasmic alterations not readily captured by the mere assessment of cell number and size. Cells that are unequivocally agreed to be ballooned are surprisingly uncommon, with only 8 cells being identified with concordance of ≥8 of 9 pathologists and one cell being unanimously considered ballooned (Figure 7).
Figure 7.
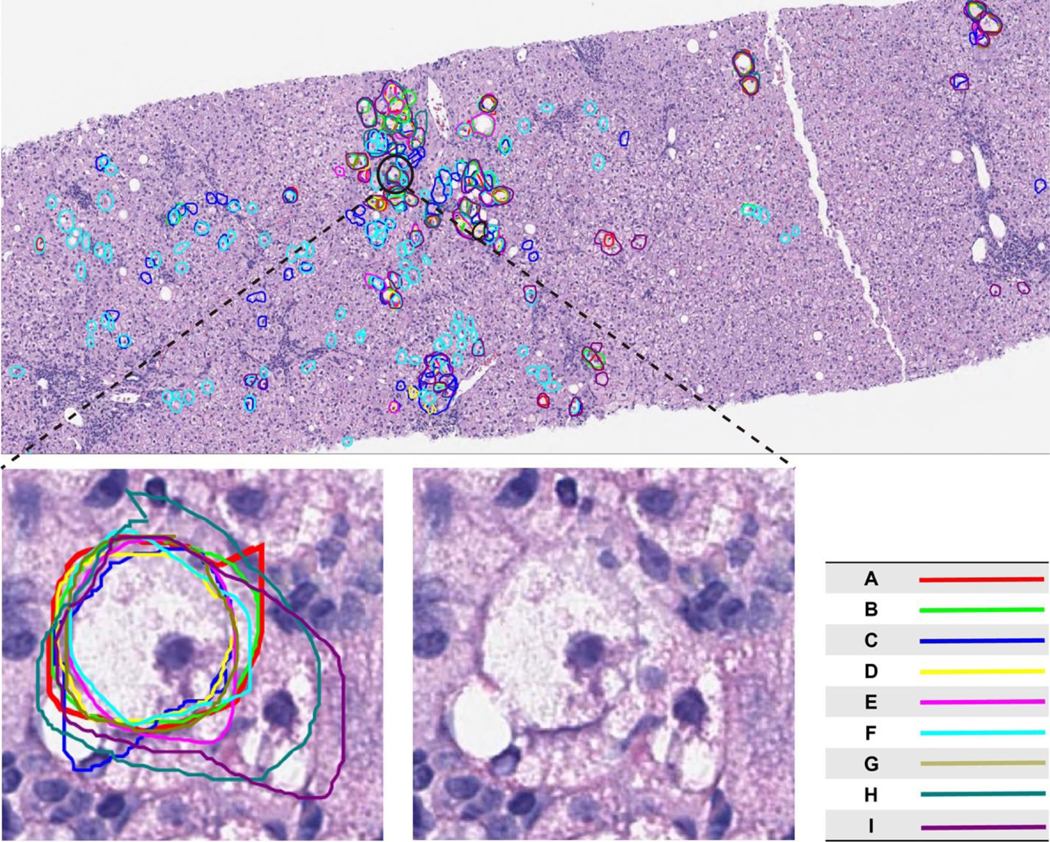
The figure shows a typical digital biopsy image used for evaluation in the study (slide #3). Lines drawn in each colour represent annotation by a different pathologist. A single hepatocyte was considered to exhibit features consistent with ballooning by all nine pathologists. The encircled ballooned hepatocyte shows features commonly described for ballooning: size greater than its neighbouring cells; flocculent cytoplasm; hyperchromatic nucleus; location near a terminal hepatic venue. This image is further magnified to demonstrate these features.
These data have important implications for drug development and the conduct of clinical trials. The magnitude of variation in the number of ballooned hepatocytes identified in any given image was sufficient to alter classification within a 3-point semi-quantitative score and so could influence eligibility decisions for trial inclusion. However, of greater importance is how this could affect trial endpoint assessment. The FDA industry guidance document explicitly defines ‘resolution of steatohepatitis’ as absent fatty liver disease or isolated or simple steatosis without steatohepatitis and a NAS score of 0–1 for inflammation, 0 for ballooning, and any value for steatosis [26]. The substantial variation in the number of ballooned cells identified, and the lack of consensus amongst the pathologists that any of the histology images were entirely ballooned-hepatocyte free (Figure 2A), implies that any trial endpoint founded on an assertion of the complete absence of ballooning (i.e. NASH CRN or SAF score zero for ballooning) is subject to substantial interobserver variation in reporting, undermining reproducibility of results based on this definition. Furthermore, our data demonstrate that there was at best only limited correlation between presence of ballooning and an overall determination of non-NASH NAFL vs NASH (Figure 5). Taken together, the degree of interobserver variation and the limited impact of this determination on non-NASH NAFL vs NASH classification suggest that there may be too great an emphasis placed on determining the presence or complete absence of ballooned hepatocytes from a given biopsy in clinical trials within the current regulatory framework.
The use of machine learning/artificial intelligence-based approaches has been proposed as a route to standardise biopsy assessment and minimise interobserver variation [16, 18]. However, as we demonstrate, the human histological reference standard is unable to produce a completely error-free classification with respect to the target condition. Although not unique to liver histopathology, such situations are methodologically challenging [32]. In the current study, we leveraged the ‘wisdom of the crowd’ [33] to train an in silico algorithm based on features detected using second harmonic generation/two-photon excitation fluorescence (SHG/TPEF) microscopy to identify ballooned hepatocytes. A reductive algorithm selected 7 parameters that assess the tissue microstructure and autofluorescent properties when the biopsy samples are irradiated with a laser [16]. As shown in Table 1, the performance of the algorithm may be adjusted according to the pre-specified concordance threshold used to train the algorithm. Selecting only ‘high concordance’ cells, in which multiple pathologists agreed on ballooning, reduced sensitivity but better controlled false positive determinations. However, this approach also limited the number of cells available for algorithm training and so a pragmatic concordance threshold of ≥5 pathologists was adopted as an exemplar. Although performance may be improved by further refinement and validation will be required before implementation, the consequent qBallooning2 algorithm is tuned to reproducibly detect a spectrum of ballooned hepatocytes based upon these SHG/TPEF parameters. Depending on the clinical context the algorithm could be calibrated differently for diagnosis or for the detection of clinically relevant temporal changes for instance in therapeutic trials. The pilot data presented here demonstrate it has the capacity to detect change in ballooning deemed relevant to identify drug-induced histological changes (Figure 7). Thus, application of artificial intelligence approaches offers a potential assistive technology that may complement human pathology where there is a need for reproducible cut-points that determine go/no-go decisions in drug development.
It is apparent that the process of developing an atlas of ballooned hepatocytes provides the opportunity for further study to elucidate additional cellular ballooning characteristics that may be more tractable for use with light microscopy, and to study the concept of change in ballooned cell burden rather than complete elimination as a potentially more viable approach for efficacy assessment. The performance of qBallooning2 as a measure of treatment response will require substantial further validation before it can be proposed as a solution to these challenges, but such validation falls outside the scope of the current manuscript.
A number of features of the study should be noted as these may be of relevance when extrapolating from these findings to other settings. Firstly, the study was undertaken using digital images as has been approved by the FDA for clinical trials in NASH and is now the case in the majority of studies. Whilst the adoption of high-resolution digital images is also becoming increasingly widespread in clinical practice and was essential to permit individual cells to be annotated by each pathologist, some of the pathologists may have been less comfortable examining digital images however individual training and a substantial practice slide-set were provided. Secondly, as the study sought to capture information on the cells that each expert pathologist identified as ballooned in independent practice, no pre-harmonisation discussions amongst the group were conducted in order to avoid introducing any bias. For the same reason, no specific guidance on how to identify ballooned cells was provided; the pathologists were instructed to identify all ballooned cells using whichever features visible on the haematoxylin and eosin-stained sections they thought appropriate. Whilst harmonisation and detailed instructions as to how to interpret features may conceivably have reduced interobserver variation, each pathologist was an independent expert in their own right and doing so would have undermined the goals of the study. It is also notable that four of the pathologists were members of the NASH CRN histopathology group and the degree of interobserver variation amongst those that were members of this long-standing collaborative team was comparable to those that were not, suggesting that further harmonisation would not have substantially reduced ballooning misclassification at the cellular level. Thirdly, we did not record how extraneous factors such as tissue and/or slide preparation quality or the premise for the study may have influenced interpretation. Some variation in staining was deliberately present in the image-set although all images met a minimum technical quality threshold.
In conclusion, we demonstrate substantial divergence in the identification of hepatocyte ballooning amongst a group of expert hepatopathologists. This appears, at least in part, to be due to differences in how subtle histopathological features are assessed by individuals and does not appear to be driven by level of experience in assessing NAFLD. Our findings have important implications for the use of ballooning as a component of treatment efficacy assessment in clinical trials, primarily because it appears that the identification of ballooning is too nuanced and subjective for its complete absence to be reliably established or adequately measured using a 3-point semiquantitative scale. In light of this, we suggest that less emphasis is placed on this single histological feature, or less evidence on absolute absence, as a marker of therapeutic efficacy. As an exemplar of how these challenges may be addressed going forward, we demonstrate that a concordance atlas may be used to train artificial intelligence/machine learning tools so that assistive technologies, whilst themselves imperfect, may standardise the quantification of histological features used to assess therapeutic efficacy.
Supplementary Material
Highlights.
Hepatocyte ballooning identification underpins regulatory-approved drug efficacy endpoints in clinical trials.
We report substantial variation in ballooned cell identification by expert hepatopathologists.
Our data suggest that ballooning is too subjective for its presence or complete absence to be unequivocally determined by pathologists as a trial endpoint.
A ‘concordance atlas’ of cells identified as ballooned by multiple pathologists can be used to train artificial intelligence (AI)-based image analysis.
AI-based approaches may provide a more reliable way to assess the range of injury recorded as hepatocyte ballooning.
Acknowledgements
This study has been supported by Histoindex Pte Ltd (DT, EC, YR); the Newcastle NIHR Biomedical Research Centre (QMA); the Intramural Research Program of the NIH, National Cancer Institute (DEK); and the LITMUS (Liver Investigation: Testing Marker Utility in Steatohepatitis) consortium funded by the Innovative Medicines Initiative (IMI2) Program of the European Union under Grant Agreement 777377; this Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA (QMA, CL, DGT, VR, SH, DT).
AJS is president of Sanyal Bio. He reports consultancy fees from Merck, Regeneron, Alnylam, Genentech, Amgen, Pfizer, Novo Nordisk, Eli Lilly, Boehringer Ingelhiem, Inventiva, Madrigal, Malinckrodt, Salix, Genfit, Hemoshear, Histoindex, Siemens, Gilead, NGM, Terns, Rivus, Endiva, 89Bio, Akero, Blade, Novartis, Axcella, Intercept; grant funding from Intercept, Gilead, Bristol Myers Squibb, merck, Pfizer, Novo Nordisk, Astra Zeneca, Boehringer Ingelhiem, Eli Lilly, Viking, Madrigal, Akero, Hanmi; stock/stock options from Genfit, Tiziana, Indalo, Exhalenz, Hemoshear; royalties from Elsevier.
AW reports fees from Clinnovate.
BNT reports consultancy fees from Histoindex.
CDG reports consultancy fees from HistoIndex, NGMBio, Cymabay and Madrigal. CDG has or has had consulting agreements with the following companies: HistoIndex, NGM, CymaBay, Madrigal and 89Bio. DGT reports consultancy for Intercept Pharmaceuticals Inc, Allergan plc, Verily Life Sciences LLC, Cirius Therapeutics Inc, Alimentiv Inc, Clinnovate Health UK Ltd, ICON Clinical Research Ltd and grants from Histoindex Pte Ltd.
MR reports consultancy fees from Alnylam, Amgen, AMRA, BMS, Boehringer Ingelheim, Centara, Coherus, Enanta, Galecto, Intercept Pharmaceuticals, Madrigal, NGM, Biopharmaceuticals, Novo Nordisk, Pfizer, Fractyl, Gelesis, Siemens, Thetis, Terns, Rivus, 3vBio (Sagimet), 89Bio and Novartis, Immuron, Merck, Taiwan J.
QMA is coordinator of the IMI2 LITMUS consortium, which is funded by the EU Horizon 2020 programme and EFPIA. This multi-stakeholder consortium includes industry partners. He reports research grant funding from Allergan/Tobira, AstraZeneca, GlaxoSmithKline, Glympse Bio, Novartis Pharma AG, Pfizer Ltd., Vertex; consultancy on behalf of Newcastle University for 89Bio, Allergan/Tobira, Altimmune, AstraZeneca, Axcella, Blade, BMS, BNN Cardio, Cirius, CymaBay, EcoR1, E3Bio, Eli Lilly & Company, Galmed, Genentech, Genfit, Gilead, Grunthal, HistoIndex, Indalo, Intercept, Inventiva, IQVIA, Janssen, Madrigal, MedImmune, Medpace, Metacrine, NGMBio, North Sea Therapeutics, Novartis, Novo Nordisk A/S, PathAI, Pfizer Ltd., Poxel, ProSciento, Raptor Pharma, Roche, Servier, Terns, The Medicines Company, Viking Therapeutics; and speaker fees from Abbott Laboratories, Allergan/Tobira, BMS, Clinical Care Options, Falk, Fishawack, Genfit SA, Gilead, Integritas Communications, Kenes, MedScape.
SAH reports grants from Akero, Axcella, Cirius, CiVi, Cymabay, Enyo, Galectin, Galmed, Genfit, Gilead Sciences, Hepion, Hightide, Intercept, Madrigal, Metacrine, NGM, Northsea, Novartis, Novo Nordisk, Poxel, Sagimet, Viking; consultancy fees from AgomAB, Akero, Alentis, Alimentiv, Altimmune, Axcella, Boston Pharma, B Riley, BVF Partners, Canfite, CiVi, Corcept, Cymabay, Echosens, Enyo, Fibronostics, Foresite, Fortress, Galectin, Genfit, Gilead, GNS, Hepion, Hightide, HistoIndex, Inipharm, Intercept, Ionis, Kowa, Madrigal, Metacrine, Microba, NGM, NorthSea, Novartis, Novo Nordisk, Nutrasource, Piper Sandler, Poxel, Prometic Pharma, Sagimet, Sonic Incytes, Terns, Viking; and stock holdings or stock options in Akero, Chronwell, Cirius, Galectin, Genfit, Hepion, HistoIndex, Metacrine, NGM, NorthSea, PathAI, Sonic Incytes. VR reports consultancy fees from Novo-Nordisk, Intercept, NGM, Theratechnologies, Galmed, Madrigal, Hanmi, Astra-Zeneca.
VT reports research grants from Gilead sciences and consulting fees from Novo-Nordisk, Intercept, NGM, Theratechnologies, Galmed, Madrigal, Hanmi, Astra-Zeneca.
Footnotes
Conflicts of Interest
DT is an employee of, and holds stock in, Histoindex. EC and RY are employees of Histoindex.
EMB has been on Advisory Board for Pfizer Ltd. And served as a consultant for Arrowhead, Cymabay, Intercept Pharmaceuticals, Medpace, Perspectum Diagnostics and Histoindex. Paid slide evaluation for Cymabay, Medpace and Perspectum Diagnostics.
The other authors report no relevant conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].EASL-EASO-EASD. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol 2016;64:1388–1402. [DOI] [PubMed] [Google Scholar]
- [2].Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328–357. [DOI] [PubMed] [Google Scholar]
- [3].Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 2013;10:330–344. [DOI] [PubMed] [Google Scholar]
- [4].Estes C, Anstee QM, Arias-Loste MT, Bantel H, Bellentani S, Caballeria J, et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016–2030. J Hepatol 2018;69:896–904. [DOI] [PubMed] [Google Scholar]
- [5].O’Hara J, Finnegan A, Dhillon H, Ruiz-Casas L, Pedra G, Franks B, et al. Cost of non-alcoholic steatohepatitis in Europe and the USA: The GAIN study. JHEP Rep 2020;2:100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rinella ME, Tacke F, Sanyal AJ, Anstee QM, participants of the A-EW. Report on the AASLD/EASL joint workshop on clinical trial endpoints in NAFLD. J Hepatol 2019;71:823–833. [DOI] [PubMed] [Google Scholar]
- [7].McPherson S, Hardy T, Henderson E, Burt AD, Day CP, Anstee QM. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: Implications for prognosis and clinical management. J Hepatol 2015;62:1148–1155. [DOI] [PubMed] [Google Scholar]
- [8].Taylor RS, Taylor RJ, Bayliss S, Hagstrom H, Nasr P, Schattenberg JM, et al. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology 2020;158:1611–1625 e1612. [DOI] [PubMed] [Google Scholar]
- [9].Food and Drug Administration (FDA). Noncirrhotic Nonalcoholic Steatohepatitis With Liver Fibrosis: Developing Drugs for Treatment Guidance for Industry. 2018. [Google Scholar]
- [10].European Medicines Agency (EMA). Draft reflection paper on regulatory requirements for the development of medicinal products for chronic non-infectious liver diseases (PBC, PSC, NASH; ). 2018. [Google Scholar]
- [11].Gramlich T, Kleiner DE, McCullough AJ, Matteoni CA, Boparai N, Younossi ZM. Pathologic features associated with fibrosis in nonalcoholic fatty liver disease. Human pathology 2004;35:196–199. [DOI] [PubMed] [Google Scholar]
- [12].Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313–1321. [DOI] [PubMed] [Google Scholar]
- [13].Bedossa P, Consortium FP. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic fatty liver disease. Hepatology 2014;60:565–575. [DOI] [PubMed] [Google Scholar]
- [14].Kleiner DE, Brunt EM, Wilson LA, Behling C, Guy C, Contos M, et al. Association of Histologic Disease Activity With Progression of Nonalcoholic Fatty Liver Disease. JAMA Netw Open 2019;2:e1912565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Davison BA, Harrison SA, Cotter G, Alkhouri N, Sanyal A, Edwards C, et al. Suboptimal reliability of liver biopsy evaluation has implications for randomized clinical trials. J Hepatol 2020;73:1322–1332. [DOI] [PubMed] [Google Scholar]
- [16].Liu F, Goh GB, Tiniakos D, Wee A, Leow WQ, Zhao JM, et al. qFIBS: An Automated Technique for Quantitative Evaluation of Fibrosis, Inflammation, Ballooning, and Steatosis in Patients With Nonalcoholic Steatohepatitis. Hepatology 2020;71:1953–1966. [DOI] [PubMed] [Google Scholar]
- [17].Wang Y, Vincent R, Yang J, Asgharpour A, Liang X, Idowu MO, et al. Dual-photon microscopy-based quantitation of fibrosis-related parameters (q-FP) to model disease progression in steatohepatitis. Hepatology 2017;65:1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Taylor-Weiner A, Pokkalla H, Han L, Jia C, Huss R, Chung C, et al. A Machine Learning Approach Enables Quantitative Measurement of Liver Histology and Disease Monitoring in NASH. Hepatology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harrison SA, Bashir MR, Guy CD, Zhou R, Moylan CA, Frias JP, et al. Resmetirom (MGL-3196) for the treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2019;394:2012–2024. [DOI] [PubMed] [Google Scholar]
- [20].Harrison SA, Gunn NT, Khazanchi A, Guy CD, Brunt EM, Mousse S, et al. A 52-Week Multi-Center Double-Blind Randomized Phase 2 Study of Seladelpar, a Potent and Selective Peroxisome Proliferator-Activated Receptor Delta (Ppar-Delta) Agonist, in Patients with Nonalcoholic Steatohepatitis (Nash). Hepatology 2020;72:1042–1043. [Google Scholar]
- [21].Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420–428. [DOI] [PubMed] [Google Scholar]
- [22].Fleiss JL. Measuring nominal scale agreement among many raters. Psychological Bulletin 1971;76:378–382. [Google Scholar]
- [23].Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–174. [PubMed] [Google Scholar]
- [24].Bedossa P, Poitou C, Veyrie N, Bouillot JL, Basdevant A, Paradis V, et al. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology 2012;56:1751–1759. [DOI] [PubMed] [Google Scholar]
- [25].Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology 1999;116:1413–1419. [DOI] [PubMed] [Google Scholar]
- [26].US Food and Drug Administration (FDA). Noncirrhotic Nonalcoholic Steatohepatitis With Liver Fibrosis: Developing Drugs for Treatment Guidance for Industry (Draft Guidance). 2018. [cited; Available from: https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM627376.pdf
- [27].Siddiqui MS, Harrison SA, Abdelmalek MF, Anstee QM, Bedossa P, Castera L, et al. Case definitions for inclusion and analysis of endpoints in clinical trials for nonalcoholic steatohepatitis through the lens of regulatory science. Hepatology 2018;67:2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guy CD, Suzuki A, Zdanowicz M, Abdelmalek MF, Burchette J, Unalp A, et al. Hedgehog pathway activation parallels histologic severity of injury and fibrosis in human nonalcoholic fatty liver disease. Hepatology 2012;55:1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lackner C, Gogg-Kamerer M, Zatloukal K, Stumptner C, Brunt EM, Denk H. Ballooned hepatocytes in steatohepatitis: the value of keratin immunohistochemistry for diagnosis. J Hepatol 2008;48:821–828. [DOI] [PubMed] [Google Scholar]
- [30].Estep M, Mehta R, Bratthauer G, Alaparthi L, Monge F, Ali S, et al. Hepatic sonic hedgehog protein expression measured by computer assisted morphometry significantly correlates with features of non-alcoholic steatohepatitis. BMC Gastroenterol 2019;19:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Caldwell S, Ikura Y, Dias D, Isomoto K, Yabu A, Moskaluk C, et al. Hepatocellular ballooning in NASH. J Hepatol 2010;53:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rutjes AW, Reitsma JB, Coomarasamy A, Khan KS, Bossuyt PM. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol Assess 2007;11:iii, ix–51. [DOI] [PubMed] [Google Scholar]
- [33].Aristotle. Politics III.1281b. Translated by Rackham H, Loeb Classical Library. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and annotated images used in this study are presented in the Supplementary Image File.