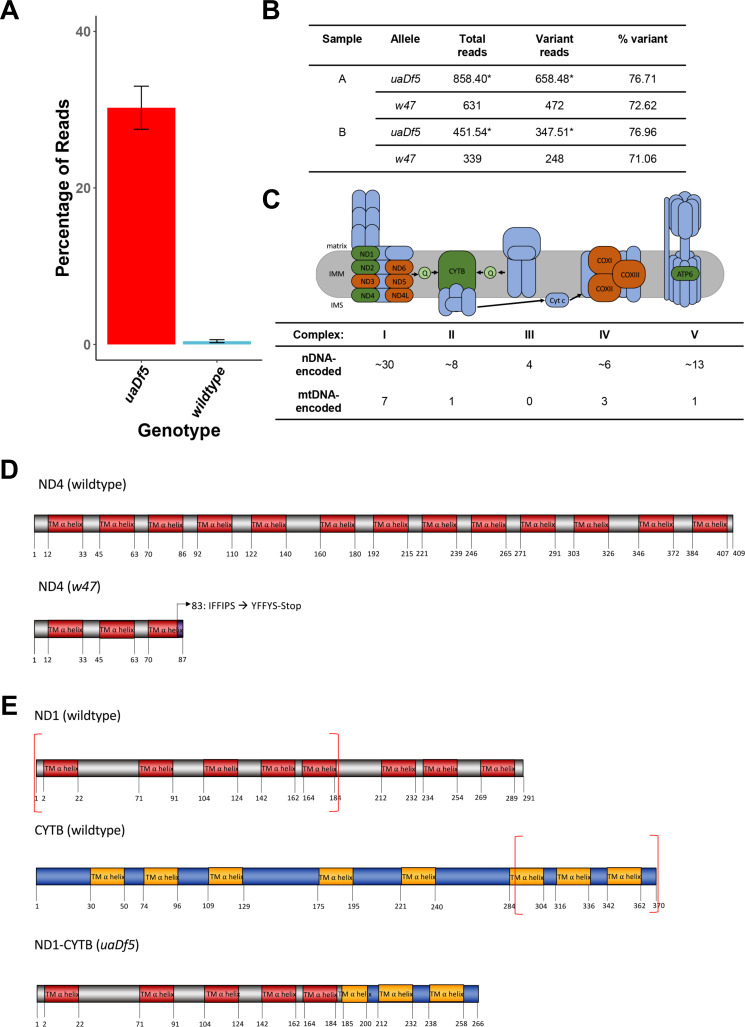
Figure 1. Analysis of the impact of mtDNAuaDf5 on fitness parameters.
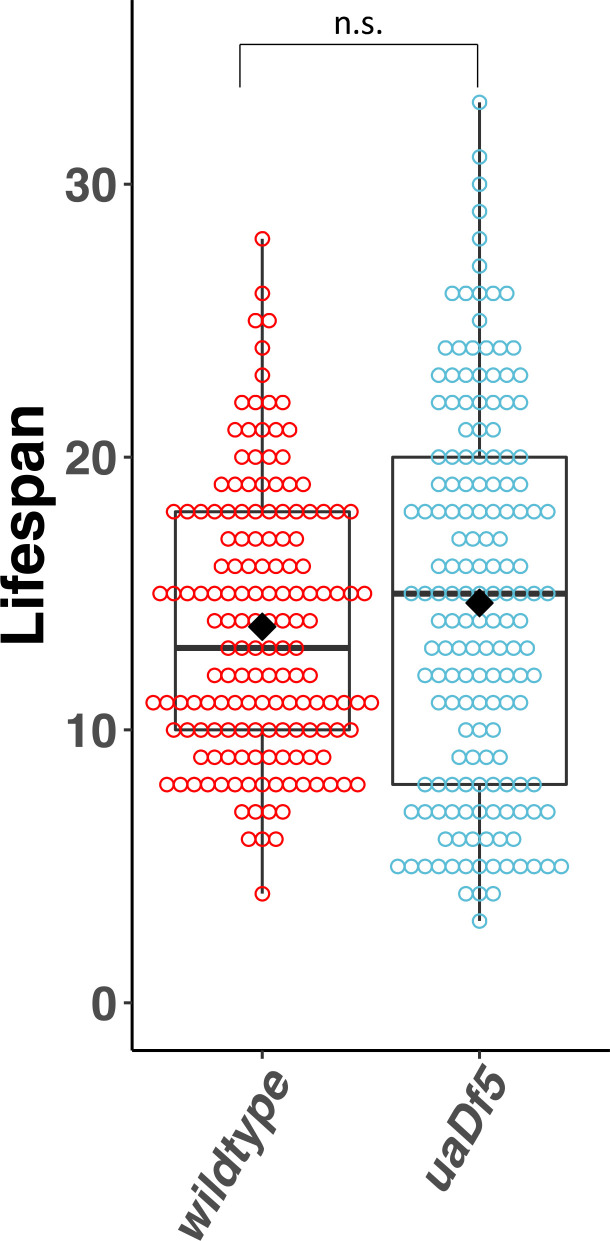
(A) Diagram of C. elegans mtDNA. Black bars with arrows indicate the locations of genes and direction of transcription. Black lines with letters indicate the locations of tRNAs. Green bars show the locations of the mtDNAuaDf5 deletion as well as the linked w47 insertion that was identified via Illumina sequencing. (B) Brood size analysis of mtDNAuaDf5 compared to laboratory wildtype N2. (C) Embryonic lethality analysis of uaDf5 compared to laboratory wildtype N2. (D) Developmental rate analysis of mtDNAuaDf5 compared to laboratory wildtype N2, counting how many hours it takes for starved L1s to reach gravidity once plated on food. (E) Developmental rate analysis of mtDNAuaDf5 compared to laboratory wildtype N2, staging worms 60 hr after synchronized, starved L1s are plated on food. (F) Survival curve analysis of mtDNAuaDf5 compared to laboratory wildtype N2, day 1 is defined as the day starved L1s are plated on food. Median lifespan and statistics are presented in Figure 1—figure supplement 2. For (B–E), box plots show median and IQR (Interquartile Range), and the diamond indicates the mean. Statistical analysis was performed using the Mann–Whitney test (***p < 0.001, n.s. not significant).