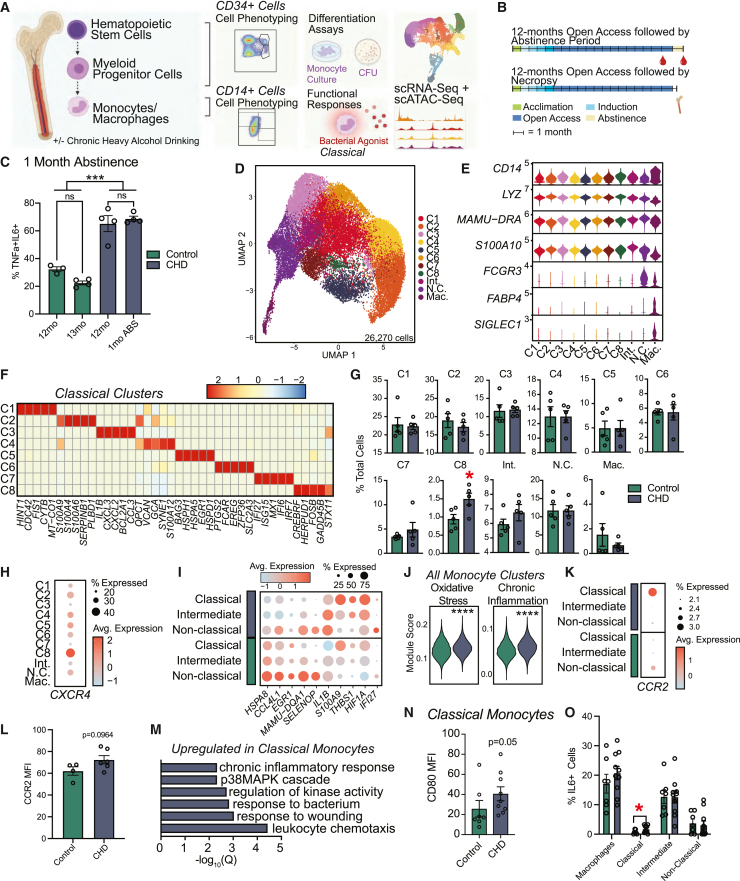
Figure 1.
Shifts in the single cell transcriptional profiles of bone marrow resident CD14+ cells with CHD
(A) Experimental design for study generated on Biorender.com.
(B) Timeline for blood collected after 1 month of abstinence (top) and bone marrow collected after 12 months of CHD (bottom).
(C) Bar plots showing the percent of TNF-α/IL-6+ monocytes measured using intracellular cytokine staining after 16 h of stimulation with bacterial TLR ligands performed on total PBMCs after 12 months of chronic drinking and 1-month abstinence for CHD animals and time-matched housing controls. Frequency of responding cells was corrected for the unstimulated condition. Error bars indicate SEM. ∗∗∗p < 0.001.
(D–M) scRNA-seq of 26,270 CD14+ bone marrow resident cells obtained from n = 3 female controls pooled (due to low cell numbers), three female CHDs pooled, four male controls, four male CHDs.
(D) UMAP clustering of cells.
(E) Violin plot showing expression of marker genes.
(F) Heatmap of average expression for highly expressed marker genes for classical monocyte clusters.
(G) Bar plots showing percentage of total cells contributing to each cluster for controls and CHDs. Error bars indicate SEM. ∗p < 0.05.
(H) Dot plot showing expression of CXCR4 across each cluster. Size of the dot represents percent of cells expressing the gene and the color is the average expression value.
(I) Dot plot showing expression of up- and downregulated DEGs with CHD common to all monocyte subsets. Size of the dot represents percent of cells expressing the gene and the color is the average expression value.
(J) Violin plots representing module scores for indicated pathways in total monocytes. Statistical significance was tested using the Mann-Whitney test.
(K) Dot plot showing gene expression of CCR2 across each monocyte subset split by CHD and control. Size of the dot represents percent of cells expressing the gene and the color is the average expression value.
(L) Bar plot of CCR2 surface protein expression MFI on total monocytes in bone marrow. Error bars indicate SEM.
(M) Bar plot representing −log10(q value) enrichment scores for genes upregulated in classical monocyte clusters with CHD.
(N) Bar plot of CD80 MFI on classical monocytes. Error bars indicate SEM.
(O) Percentage of IL-6+ cells within each monocyte and macrophage population in bone marrow following stimulation with a bacterial TLR cocktail corrected for the unstimulated condition. Error bars indicate SEM. Statistical significance was tested by t test with Welch’s correction where ∗p < 0.05. See also Figures S1 and S2.