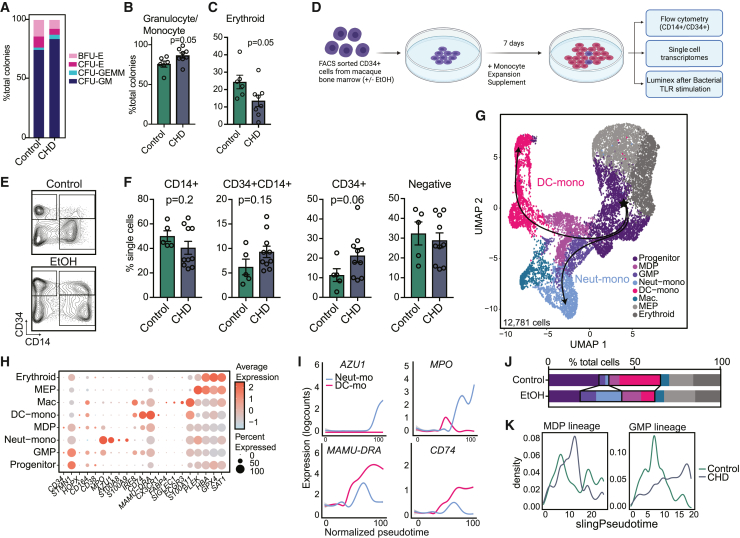
Figure 2.
CHD alters CD34+ progenitor cell differentiation to monocytes
(A) Stacked bar plot showing percentages of indicated colonies.
(B and C) Bar plots showing the percentage of granulocyte/monocyte (B) and erythroid (C) colonies from total colonies across control and CHD groups. Error bars indicate SEM.
(D) Experimental design created on Biorender.com. Sorted CD34+ cells from control and CHD macaque bone marrow were cultured in monocyte differentiation media supplement for 7 days.
(E) Example gating strategy showing CD34+ versus CD14+ cells.
(F) Bar plots showing quantification of the culture output by flow cytometry. Error bars indicate SEM.
(G–K) scRNA-seq of 12,781 cells from pooled controls (n = 2 male + 2 female) and pooled CHD (n = 2 male + 2 female).
(G) UMAP with overlapped Slingshot pseudotime trajectory lines.
(H) Dot plot showing expression of marker genes. Size of the dot represents percent of cells expressing the gene and the color represents an average expression value.
(I) Log expression of AZU1, MPO, MAMU-DRA, and CD74 plotted for each cell across the indicated scaled Slingshot pseudotime trajectory (trendline shown).
(J) Bar plots showing representative percentages of each cluster across control and CHD groups.
(K) Cell density plots for MDP (left) and GMP (right) lineage determined by Slingshot. Statistical significance was tested by t test with Welch’s correction where ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. See also Figure S3.