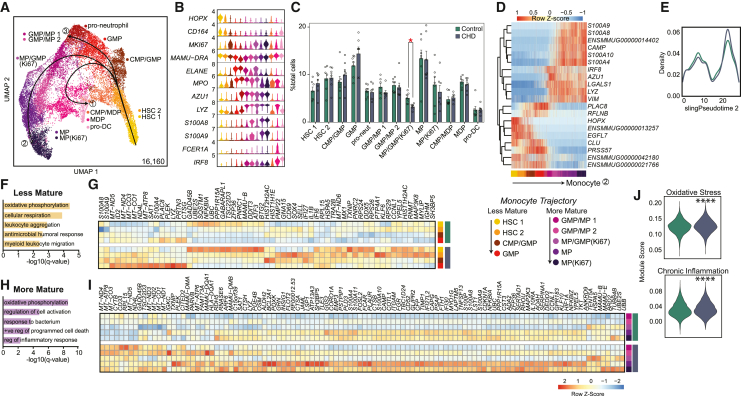
Figure 4.
CHD CD34+ bone marrow myeloid progenitor single cell transcriptional profiles
scRNA-seq of 16,610 cells from three female and four male controls, and three female and four male CHD animals.
(A) UMAP with Slingshot trajectory lines.
(B) Stacked violin plot of marker gene expression.
(C) Cluster percentages between CHD and control groups. Compared using two-way ANOVA with correction for multiple comparisons. Error bars indicate SEM.
(D) Heatmap of genes that dictate the monocyte lineage trajectory.
(E) Cell density plot for control and CHD groups across the monocyte trajectory lineage.
(F and H) Functional enrichment terms for DEGs upregulated with CHD from less (F) and more (H) mature clusters split by CHD and control groups.
(G and I) Heatmap of averaged gene expression for upregulated DEGs from less (G) and more (I) mature clusters split by CHD and control groups.
(J) Violin plots for indicated module scores across groups. Statistical analysis was performed by the Mann-Whitney test. Unless indicated, statistical significance was tested by t test with Welch’s correction. ∗p < 0.05, ∗∗∗∗p < 0.0001. See also Figures S4 and S5.