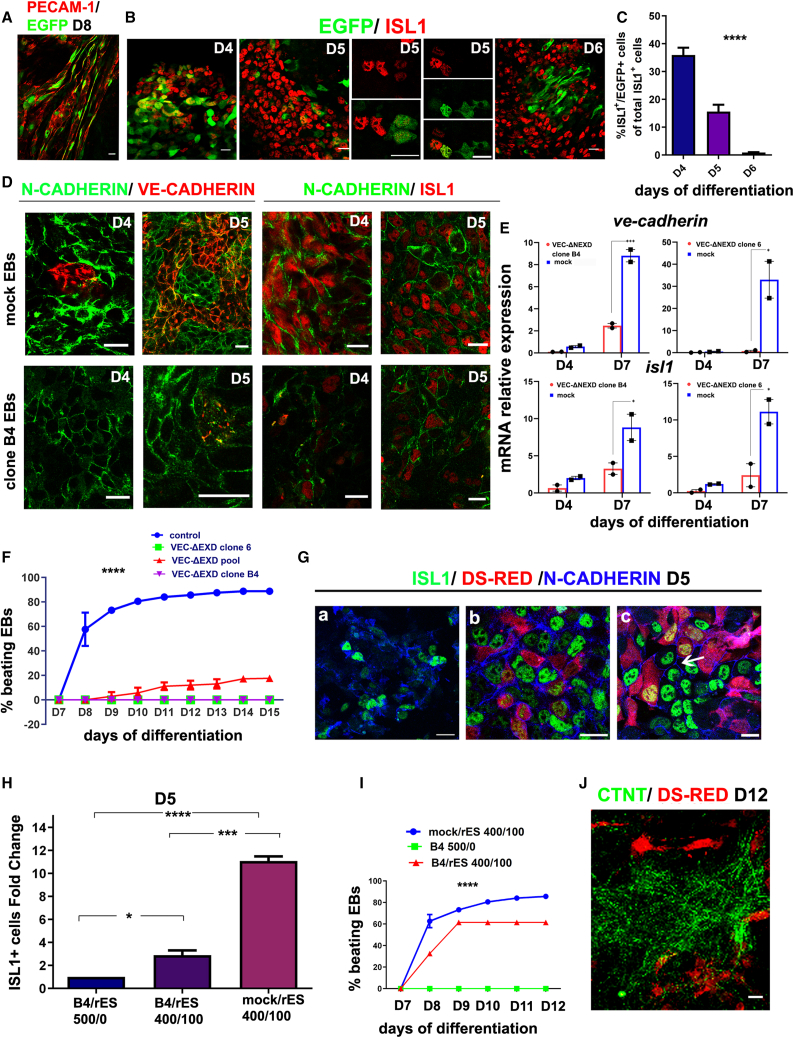
Figure 4.
Inhibition of classical cadherins in Pvec+ cells severely affected cardiac differentiation of ESCs
(A) Pvec activity is restricted to endothelial cells on day 8, as indicated by PECAM-1/EGFP co-expression.
(B) ISL1+ cells express EGFP in EBs on days 4–6. Note that Pvec activity was high in low ISL1 expressors and, conversely, on day 5, implying that Pvec is activated transiently during ISL1+ cell specification.
(C) Statistical analysis of ISL1+/EGFP+ cells on days 4–6. Approximately 1,000 ISL1+ cells were counted per experiment. Three independent experiments were performed. ∗∗∗∗p < 0.0001.
(D) VE- and N-CADHERIN AJs in clone B4 and mock EBs on days 4 and 5. Note the absence or disruption of VE-CADHERIN, partial disruption of N-CADHERIN, and significant decrease of ISL1+ cells in clone B4 EBs compared with mock.
(E) Endogenous ve-cadherin and isl1 mRNA levels in clones B4 and 6 and mock quantified by real-time qPCR during the indicated differentiation days (n = 2).
(F) Statistical analysis of beating activity in clones B4, 6, pool, and mock during differentiation days 7–15 (an EB was considered beating when it contained one or more beating areas). The numbers of EBs counted were as follows: mock, 260; clone B4, 185; clone 6, 210; pool, 217 (three independent biological experiments, n = 3). ∗∗∗∗p < 0.0001.
(G) N-CADHERIN, dsRed, and ISL1 triple staining in EBs from clone B4 (a), mock/rES (b), and B4/rES(c) on day 5. EBs were dissociated under mild trypsinization on day 3, and cells were left to attach on fibronectin and grow as a monolayer. Note that the number of ISL1+ cells in hybrid B4/rES EBs are substantially increased compared with B4 and form extensive AJs comparable with mock/rES (arrow).
(H) ISL1+ cell fold change between mutant B4 and hybrid B4/rES and mock/rES. The number of ISL1+ cells counted in each case was derived specifically from mutant and mock ESCs and not from rES. For this experiment, 10 random fields were photographed, and approximately 400 cells per field were counted in three independent experiments. Statistical analysis shows the fold change of ISL1+ cells compared with B4 values set to 1. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(I) Statistical analysis of beating activity in hybrid EBs during differentiation days 7–12 (three independent biological experiments, n = 3). ∗∗∗p < 0.001.
(J) Rescue of cTNT expression in hybrid EBs (B4/rES) on day 12 (maximum projection of confocal image series).
Scale bars, 20 μm. In (C), the clone B4 EB day 5 N/VE-CADHERIN scale bar represents 40 μm. Statistical analysis details are described in the supplemental information. All experiments were performed at least three times, and representative data are shown.