Abstract
Nuclear receptor subfamily 2 group F member 2 (NR2F2 or COUP-TF2) encodes a transcription factor which is expressed at high levels during mammalian development. Rare heterozygous Mendelian variants in NR2F2 were initially identified in individuals with congenital heart disease (CHD), then subsequently in cohorts of congenital diaphragmatic hernia (CDH) and 46,XX ovotesticular disorders/differences of sexual development (DSD); however, the phenotypic spectrum associated with pathogenic variants in NR2F2 remains poorly characterized. Currently, less than 40 individuals with heterozygous pathogenic variants in NR2F2 have been reported. Here, we review the clinical and molecular details of 17 previously unreported individuals with rare heterozygous NR2F2 variants, the majority of which were de novo. Clinical features were variable, including intrauterine growth restriction (IUGR), CHD, CDH, genital anomalies, DSD, developmental delays, hypotonia, feeding difficulties, failure to thrive, congenital and acquired microcephaly, dysmorphic facial features, renal failure, hearing loss, strabismus, asplenia, and vascular malformations, thus expanding the phenotypic spectrum associated with NR2F2 variants. The variants seen were predicted loss of function, including a nonsense variant inherited from a mildly affected mosaic mother, missense and a large deletion including the NR2F2 gene. Our study presents evidence for rare, heterozygous NR2F2 variants causing a highly variable syndrome of congenital anomalies, commonly associated with heart defects, developmental delays/intellectual disability, dysmorphic features, feeding difficulties, hypotonia, and genital anomalies. Based on the new and previous cases, we provide clinical recommendations for evaluating individuals diagnosed with an NR2F2-associated disorder.
Subject terms: Neurodevelopmental disorders, Clinical genetics
Introduction
Nuclear receptor subfamily 2 group F member 2 (NR2F2, also known as COUP-TF2) gene encodes a member of the nuclear receptor superfamily of ligand-activated transcriptional factors involved in several developmental and cellular processes. NR2F2 is an orphan receptor whose ligand is yet to be identified. Similar to the other nuclear receptors, NR2F2 protein has three main domains: an N-terminal activation binding motif (1-78aa), a DNA-binding domain (79-151aa), and a C-terminal ligand-binding domain (177-411aa) separated by the hinge region [1, 2].
The spatiotemporal expression pattern of NR2F2 during mammalian development has been studied in mouse models [1, 3, 4]. Nr2f2 expression is observed between E8.5 and E13.5 in the sinus venosus, umbilical veins, heart, atrium, branchial arches, developing hindbrain, neuroectoderm of anterior midbrain, somites, otocyst, the periocular mesenchyme, optic stalk, olfactory placode, developing testes, the genital tubercle, mesenchyme of the kidney, and the adrenal cortex [1, 4]. High expression of NR2F2 is observed in the mesenchymal component of several organs during development and organogenesis [1, 3].
Homozygous deletion of Nr2f2 is embryonic lethal in mice. The Nr2f2−/− embryos show growth restriction, edematous cysts, severe hemorrhage in the brain and heart, and die around 10 days of gestation [4]. Heterozygous Nr2f2 knockout mice are smaller than the wild-type mice and show poor postnatal viability. Furthermore, Nr2f2 knockout mice show defects in sinus venosus development, dysplastic anterior and posterior cardinal veins, atrial malformations, and angiogenesis defects [4]. A recent study showed that female mouse embryos lacking Nr2f2 in the Wolffian duct mesenchyme develop as intersex, that is, having both female and male reproductive tracts [5]. Other Nr2f2 mouse models showed developmental anomalies of the female reproductive system [6–8], anteroposterior patterning of the stomach [9], diaphragm [10], lymphangiogenesis, adipogenesis, limb, eye, and cerebellum [3].
In humans, NR2F2 is extremely intolerant to loss‐of‐function (LoF) variants (pLI = 0.99 in Genome Aggregation Database (gnomAD) v2.1.1) [11, 12]. NR2F2 heterozygous LoF variants have been associated with congenital malformations, including CHD, CDH, and DSD (Supplementary Table 1) [1, 13–24]. However, most of these studies were done using disease-specific cohorts and thus have limited clinical description, especially about the other affected organ systems.
Al Turki et al. (2014) described eight individuals with CHD who had rare LoF, missense, and splice variants, and a balanced translocation in NR2F2 (Supplementary Table 1) [1]. Subsequently, NR2F2 rare variants were reported in other CHD cohorts [14–17, 22]. High et al. (2016) identified two individuals with CDH and heterozygous LoF variants in NR2F2 [24], other CDH-cohort studies have also identified rare NR2F2 LoF variants and a splice variant [15, 19, 21, 24, 25]. In a cohort of 46,XX SRY-negative ovotesticular DSD cases, Bashamboo et al. (2018) found NR2F2 LoF variants in three individuals with syndromic clinical features including CHD, CDH and dysmorphic facial features of blepharophimosis, ptosis, and epicanthus inversus syndrome (BPES) [20]. Recently, a rare NR2F2 de novo missense variant was reported in an individual with 46,XY DSD, micropenis and hypospadias [26].
Two case reports have described small chromosome 15q26.2 deletions (<5Mb) encompassing NR2F2: a de novo mosaic 1.7Mb deletion in a fetus with CDH and coarctation of the aorta, and a de novo 3Mb heterozygous deletion in a 46,XX individual reared as male with ovotesticular DSD, dysmorphic features, sinus bradycardia, low weight, blepharophimosis, and ptosis (Supplementary Table 1) [27, 28]. Larger deletions (>5Mb) involving NR2F2 along with other genes [29–34] and a 15q26.2 deletion including NR2F2 but without precise molecular breakpoints have also been reported [35].
Here, we describe 17 previously unreported individuals with varied phenotypes, including CHD, CDH, and other affected organ systems including vascular malformations, who have rare heterozygous, mostly de novo variants in NR2F2. We review and compare the clinical and molecular data of these new cases with previously reported individuals, to delineate and expand the phenotypic and genotypic spectrum of NR2F2-associated disorders.
Materials and methods
Trio whole exome sequencing (WES) was performed for the cases, except for case 8 as the father’s testing was pending and case 12, where chromosomal microarray analysis was performed. Detailed methods are provided in the Supplementary information.
Results
Molecular findings
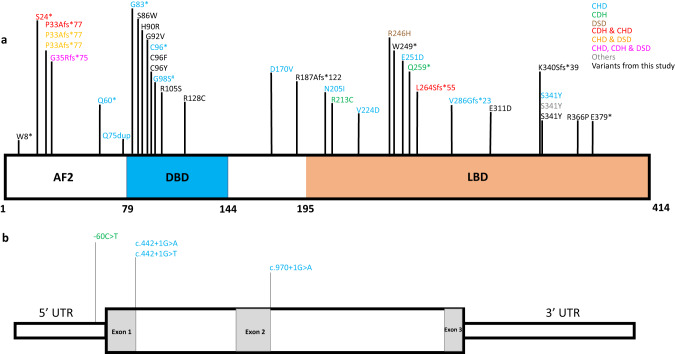
Fifteen of the seventeen newly reported individuals in our cohort had de novo, rare, heterozygous variants in NR2F2 (NM_021005.4; NP_066285.1), and one (individual 5-1) had a variant inherited from a mosaic, mildly affected mother (individual 5-2, 34% variant allelic fraction, Supplementary Table 2 and Fig. 1). The inheritance for the remaining two individuals was unknown. Sixteen of the seventeen individuals had a heterozygous NR2F2 single nucleotide variant (SNV) whereas one case (individual 12) had a large deletion. Ten of the fifteen unique NR2F2 SNVs were rare missense variants; the remaining five were predicted LoF variants. All NR2F2 variants identified in this study were absent in gnomAD (v2.1.1 and v3) and TOPMed population databases (see Table 1 for details on all variants). The distribution of NR2F2 variants from our study and previously reported cases on the encoded protein and the gene are shown in Fig. 1 and Supplementary Fig. 1.
Fig. 1. Variants in NR2F2.
a Distribution of non-synonymous variants on the NR2F2 protein (NM_021005.4, NP_066285.1). The variants identified in this study are shown in black font, and the previously published variants (except one which is marked with #) are in colored font as per the reported phenotype/clinical features. The location of the functional domains in NR2F2 protein were adapted from Wang et al. 2019 [18]. b Schematic representation of the coding exons, the splice variants, and the 5’ UTR variant reported in the NR2F2 gene (NM_021005.4). #We also reviewed whole exome sequencing data from the Pediatric Cardiac Genomics Consortium (PCGC) cohort [37, 38], which consists of individuals with cardiac defects and identified one additional case with a rare de novo NR2F2 variant (p.(Gly98Ser), Supplementary Table 4). The cardiac phenotypes, in this case, were secundum atrial septal defect, left aortic arch with normal branching pattern. This case is not included in the current cohort but has been shown in Fig. 1. CHD congenital heart defect; CDH congenital diaphragmatic hernia; DSD disorders of sexual development including 46,XX DSD and 46,XY DSD; “Others” phenotype described in Arsov et al. 2021 [14] includes asplenia. Note that though many of the previously reported affected individuals were ascertained using a disease specific disease cohort, there might be other clinical manifestations which may or may not have been described [1, 14–27] and [Supplementary Table 1].
Table 1.
Molecular details of the NR2F2 variants seen in this study along with in silico predictions for the missense variants (NM_021005.4).
| Case | Chr-position-ref-alt (hg19) | HGVS cDNA | HGVS protein | Score | rs IDs | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | Provean | REVEL | MetaSVM | CADD | GERP | |||||
| 1 | chr15-96877795-G-C | c.933G>C | p.Glu311Asp | 0 | −2.6 | P (0.822) | 1.0915 | 26.9 | 5.2699 | |
| 2 | chr15-96875621-G-T | c.287G>T | p.Cys96Phe | 0.001 | −8.48 | P (0.9599) | 0.8745 | 32 | 4.61 | |
| 3 | chr15-96875591-C-G | c.257C>G | p.Ser86Trp | 0 | −5.51 | P (0.8569) | 1.0866 | 29.9 | 4.7199 | |
| 4 | chr15-96880621-GA-G | c.1019del | p.Lys340SerfsTer39 | _ | _ | _ | _ | _ | 5.12 | |
| 5 (1 & 2) | chr15-96877608-G-A | c.746G>A | p.Trp249Ter | _ | _ | _ | _ | 39 | 5.09 | rs1567138573 |
| 6 | chr15-96877420-G-GG | c.558dup | p.Arg187AlafsTer122 | _ | _ | _ | _ | _ | 5.05 | |
| 7 | chr15-96875603-A-G | c.269A>G | p.His90Arg | 0.001 | −6.18 | P (0.985) | 1.0847 | 27.3 | 4.7199 | |
| 8 | chr15-96875621-G-A | c.287G>A | p.Cys96Tyr | 0.001 | −8.5 | P (0.97) | 0.8907 | 31 | 4.61 | |
| 9 | chr15-96880628-C-A | c.1022C>A | p.Ser341Tyr | 0.006 | −3.17 | P (0.9169) | 1.0166 | 30 | 5.15 | rs587777371 |
| 10 | chr15-96875647-C-A | c.313C>A | p.Arg105Ser | 0 | −4.65 | P (0.9319) | 0.967 | 28.7 | 4.61 | |
| 11 | chr15-96875716-C-T | c.382C>T | p.Arg128Cys | 0 | −6.79 | P (0.976) | 0.9975 | 33 | 4.92 | rs1596425042 |
| 12 | arr[GRCh37] 15q26.2q26.3(96755103_98628389)x1 | _ | _ | _ | _ | _ | _ | _ | _ | |
| 13 | chr15-96880740-A-ATA | c.1133_1134dup | p.Glu379Ter | _ | _ | _ | _ | _ | 5.44 | |
| 14 | chr15-96875609-G-T | c.275G>T | p.Gly92Val | 0.001 | −6.9 | P (0.9729) | 1.0429 | 31 | 4.61 | |
| 15 | chr15-96875357-G-A | c.23G>A | p.Trp8Ter | _ | _ | _ | _ | 37 | 4.5599 | |
| 16 | chr15-96880703-G-C | c.1097G>C | p.Arg366Pro | 0 | _ | P(0.951) | _ | 32 | 5.44 | |
For the variants when in silico prediction scores were available, they were Damaging (SIFT, Provean, MetaSVM) or Pathogenic (REVEL). The listed single nucleotide variants were not present in gnomAD v2, v3 and TOPMED population databases. Some of the listed variants were submitted to ClinVar by the laboratories which performed the exome sequencing.
CADD scores were accessed from https://cadd.gs.washington.edu/snv.
Bolded variants are predicted to be protein terminating variants.
Notably, two predicted LoF variants (p.Lys340SerfsTer39, individual 4; p.Glu379Ter, individual 13) map to the last coding exon of NR2F2 and may escape nonsense-mediated decay; no other downstream truncating variants are reported in the affected individuals. These truncations delete a part of the ligand binding domain; however, whether they lead to the loss of NR2F2 function is currently unknown.
We also reviewed whole exome sequencing data from the Pediatric Cardiac Genomics Consortium (PCGC) cohort [36, 37], which consists of individuals with cardiac defects and identified one additional case with a rare de novo NR2F2 variant (p.Gly98Ser). This case is not included in the current cohort, but has been shown in Fig. 1.
In one individual (individual 12) chromosomal microarray identified a novel, large de novo deletion (~1.87Mb, arr[GRCh37] 15q26.2q26.3(96755103_98628389)x1 at chromosome 15q26.2 encompassing NR2F2-AS1, NR2F2, SPATA8-AS1, LINC02254, LINC00923, ARRDC4, and LINC01582 genes (Supplementary Fig. 1). This deletion results in the loss of the entire NR2F2 gene and this loss is not seen in the population databases (Database of Genomic Variants (DGV), gnomAD SVs v2.1). ARRDC4, the only other protein-coding gene in this deleted region, is not constrained for LoF variants (pLI = 0, gnomAD v2) and currently has no known disease associations. Therefore, the clinical features observed in this individual were ascribed to the single copy loss of NR2F2.
Clinical findings
We describe 16 unrelated affected individuals and a mildly affected mosaic mother of one of the individuals with heterozygous variants in NR2F2. Clinical case reports for the 17 individuals are described in the Supplementary Information. These individuals had variable clinical features, including developmental delays/intellectual disability, CHDs, dysmorphic facial features, feeding difficulties, hearing impairment, hypotonia, genital anomalies, renal abnormalities, CDH, vascular malformations and prenatal findings, including CHD, IUGR, increased nuchal translucency, and single umbilical artery. The clinical features of these individuals are described in Supplementary Table 2 and the frequencies of the phenotypes observed in our cohort are listed in Table 2. The commonly observed clinical phenotypes in multiple individuals with NR2F2 variants in our cohort are described below.
Table 2.
Frequency of clinical features in our cohort.
| Clinical Features | Frequency (%) |
|---|---|
| Prenatal findings | |
| IUGR/low birth weight | 4/17 (23.5%) |
| Increased nuchal translucency | 3/17 (17.6%) |
| Microcephaly | 1/17 (5.9%) |
| CHD | 5/17 (29.4%) |
| Oligohydramnios | 2/17 (11.8%) |
| Single umbilical artery | 3/17 (17.6%) |
| Postnatal findings | |
| Microcephaly (at last examination) | 6/14 (42.9%) |
| Dysmorphic features | 16/17 (94.1%) |
| CHD | 17/17(100%) |
| CDH | 2/17 (11.8%) |
| DD/ID | 14/14 (100%) |
| Hypotonia | 10/17 (58.8%) |
| Feeding difficulties/NGT dependence | 14/17 (82.4%) |
| Seizures/staring spells/EEG abnormalities | 3/17 (17.6%) |
| Hearing impairment | 5/14 (35.7%) |
| Strabismus | 6/15 (40%) |
| Asplenia | 2/17 (11.8%) |
| Clinodactyly | 5/17 (29.4%) |
| Genital anomalies | 8/17 (47.1%) |
| Renal anomalies/issues | 6/17 (35.3%) |
| Vascular anomalies | 2/17 (11.8%) |
Prenatal findings
CHDs were detected prenatally in four of seventeen (23.5%) individuals whereas eight of the remaining individuals (47%) had prenatal findings other than CHD, including IUGR, increased nuchal translucency, single umbilical artery, oligohydramnios, hepatic vascular malformation and CDH (Supplementary Table 2).
Postnatal findings
Feeding difficulties
Fourteen of seventeen (82%) individuals had feeding difficulties and/ most with a history of nasogastric tube dependence (13/14).
Developmental delays/Intellectual disability
All 14 individuals for whom data were available had motor delays and thirteen of them also had speech delays. Delays were noted for the age at sitting, walking, and utterance of first words.
Hypotonia was observed in 59% (10/17) of the cohort. Two of the thirteen individuals (15%) for whom brain imaging data were available, showed periventricular white matter abnormalities.
CHD
All individuals (17/17) in our cohort had CHD. The commonly seen anomalies included atrial septal defect (8/17), coarctation of the aorta (8/17), ventricular septal defect (5/17), aortic arch anomalies (3/17), atrioventricular septal defect (2/17), dilated right ventricle (2/17) and pulmonary hypertension (2/17).
CDH
CDH was observed in two of seventeen (12%) individuals, and both died within a few days after birth.
Dysmorphic features
Characteristic facial features were observed in all individuals except for the mosaic mother of individual 5 (16/17; 94%). The commonly observed anomalies (seen in ≥3 affected individuals) included upslanted or short palpebral fissures, hypertelorism, low-set and/or dysplastic ears, full cheeks, and retrognathia or micrognathia (Supplementary Tables 2, 4 and Supplementary Fig. 2). Other facial features which were seen in any 2 affected individuals were microcephaly, bilateral epicanthus, deeply set eyes, long eyelashes, long philtrum, facial asymmetry, dysplastic ears, and short neck. Some of the less common features included synophrys, highly arched eyebrows, ptosis, down turned corners of mouth, and tapered finger among others (Supplementary Tables 2, 4).
Hearing loss
Five of fifteen (33%) individuals had hearing loss, which was unilateral in four cases. At least one individual needed hearing aids and another individual had conductive hearing loss.
Strabismus
Divergent strabismus was seen in 6 of 15 (40%) individuals.
Genital anomalies & DSD
Six of seven 46,XY males (86%) in the cohort had cryptorchidism. Out of the nine 46,XX females, one had labial hypertrophy.
One of the 46,XX individuals was diagnosed with DSD (individual 15, SRY negative) and had a de novo NR2F2 LoF variant (c.23G>A, p.(Trp8Ter)). Prenatally, aberrant right subclavian artery, IUGR, and oligohydramnios were detected in this individual and amniocentesis revealed a 46,XX karyotype. At birth, male external genitalia with perineal hypospadias, pigmented bifid scrotum with penoscrotal transposition and inguinal gonads were observed. There was no history of maternal virilization during the pregnancy. This 46,XX karyotype individual was diagnosed with DSD, severe aortic coarctation, and aberrant right subclavian artery. The abdominal ultrasound showed the presence of both testes, and a testis biopsy revealed testicular parenchyma comprising seminiferous ducts made up of Sertoli cells without the presence of spermatogonia, and the presence of Leydig cells in the testicular interstitium. Subsequently, bilateral orchidopexy, urethroplasty, penoscrotal transposition correction, and scrotal closure were performed.
Renal anomalies
Six of seventeen (35%) individuals had renal findings including renal failure (2/17), renal hypoplasia (1/17), and multicystic dysplastic kidneys (1/17). One of the individuals who had renal hypoplasia and congenital renal failure underwent a renal transplant. Nephrocalcinosis was also noted in one person.
Asplenia
Asplenia was noted in two of seventeen (12%) individuals.
Vascular malformations
Vascular malformations were seen in two of seventeen (12%) individuals, including one with a hepatic vascular malformation and another with vascular lesions on skin.
Skeletal abnormalities
Five out of seventeen (29%) individuals had 5th finger clinodactyly; talipes and clawing of toes were each noted in two individuals, and other skeletal findings were seen in seven out of seventeen individuals (41%; Supplementary Table 2).
Discussion
In this study, we describe 17 previously unreported individuals with rare, potentially disease causing variants in NR2F2 and provide a comprehensive delineation of the clinical and molecular features of the associated syndrome (Tables 1 and 2, Fig. 1, Supplementary Figs. 3 and 4 and Supplementary Tables 2 and 3). The clinical and molecular details of all the previously reported cases with rare heterozygous NR2F2 variants are summarized in Supplementary Table 1.
Molecular spectrum
Fourteen of the fifteen NR2F2 SNVs detected in our cohort are novel variants and have not been previously reported. Nine individuals described in our cohort carry novel rare missense NR2F2 variants. The missense variant seen in the Pediatric Cardiac Genomics Consortium case (p.(Gly98Ser)) is also not reported before [36, 37]. An additional six missense variants have been previously reported in affected individuals, one of which is recurrent in our cohort (p.(Ser341Tyr); Fig. 1 and Supplementary Table 1). These variants localize to the two conserved functional domains of NR2F2 protein, the DNA-binding domain and the ligand-binding domain (Fig. 1 and Supplementary Figs. 3 and 4).
Al Turki et al. [1] used an in vitro luciferase assay in HEK293 cells to show the functional effects of NR2F2 missense variants found in the CHD cohorts (p.(Asp170Val), p.(Asn205Ile), p.(Glu251Asp), p.(Ser341Tyr), and p.(Ala412Ser)). Luciferase assays using transfected expression plasmids of the NR2F2 variants showed a significant change in the transcriptional activity when compared with wild type NR2F2 plasmid; the direction of this effect (increased or decreased transcriptional activity) was dependent on the promoter context, that is, NGFI-A or APOB promoters which are known direct targets for NR2F2 binding [1]. These results along with the presence of NR2F2 LoF variants in the affected individuals suggest that the global mis-regulation of transcriptional activities of NR2F2 targets due to haploinsufficiency or LoF or possibly gain-of-function variants in NR2F2 during development, may lead to congenital anomalies in multiple organ systems.
Most of the reported NR2F2 missense variants, however, have not been functionally characterized. Notably, NR2F2 is constrained for missense changes (missense Z score = 3.6, gnomADv2.1.1). The pathogenicity of the NR2F2 missense variants detected in the affected individuals is supported by their absence in the population databases, the de novo status, and the pathogenic in silico predictions of their functional impact. In the future, assessing the effect of the NR2F2 missense variants using in vitro luciferase assays for the transcriptional activity or in vivo chromatin immune-precipitation (ChIP) experiments using patient-derived cells to assess the changes in levels of NR2F2 protein binding to target genomic regions, such as gene promoters or regulatory regions, will further elucidate the mechanism of these missense alterations.
Clinical features
The most common clinical features observed in our cohort included developmental delays/intellectual disability (100%), CHD (100%), dysmorphic features (94.1%), feeding difficulties or feeding tube dependence (82.4%), hypotonia (58.8%) and cryptorchidism in 46,XY males (86%, Table 2 and Supplementary Table 3).
All individuals had developmental delays, specifically, language was delayed in all individuals, with a wide range for first words (11 months to ‘not yet achieved’ at 8 years). Motor milestones were also delayed, with walking achieved at 2–3 years in some cases, but ‘not yet achieved’ at 8 years in others.
All the affected individuals reported here had CHD and extensive additional medical and developmental issues. The types of heart defects observed were similar to those reported previously, with the majority of individuals having either an atrial or ventricular septal defect or coarctation of the aorta. Three individuals had aortic arch anomalies and two individuals had atrioventricular septal defects. Other less common cardiac defects were valvular and supravalvular pulmonic stenosis, aberrant left subclavian artery, tricuspid regurgitation and bicuspid aortic valve. Two individuals had pulmonary hypertension and four had cardiac arrhythmias. Interestingly, a de novo NR2F2 variant p.(Ser341Tyr) was recently reported in a male with developmental delays, asplenia, frequent respiratory infections, dysmorphic facial features, bilateral cryptorchidism, and glandular hypospadias but with no associated CHD, CDH or DSD (Supplementary Table 1) [13]. Echocardiograms and abdominal ultrasound ruled out the presence of CHD and CDH in this individual.
Nearly all individuals in our cohort had dysmorphic facial features, most common being upslanted or short palpebral fissures, micrognathia or retrognathia, low-set or dysplastic ears, hypertelorism, and full cheeks. Other features seen were microcephaly, bilateral epicanthus, long eyelashes, long philtrum, facial asymmetry and short neck. Dysmorphic facial features seen in our cohort and in previously reported individuals with NR2F2 pathogenic variants [16, 18, 20]; [Supplementary Table 1] overlap with the findings seen in the BPES which is caused by heterozygous loss of function variants in FOXL2 gene. The shared BPES phenotype in affected individuals with FOXL2/NR2F2 pathogenic variants suggests, these genes might function together in the developmental pathways leading to eyelid formation. Similarly, both these transcription factors are known to function in mammalian ovarian development and are potential “pro-ovary/anti-testis” genes [20, 38].
Other common findings include feeding difficulties, which required requiring gastric tube due to growth issues. Clinical features that have not been commonly associated with NR2F2 variants in previously published cases, were seen in our cohort. Five individuals had hypoplastic or dysplastic kidneys, including two with subsequent chronic renal failure, five had hearing impairment, six had strabismus, and three individuals had sleep apnea or breath holding. Laryngomalacia and accessory spleen were also seen in two individuals each (Supplementary Table 2).
Two related individuals (5-1 and 5-2) had asplenia or polysplenia, a phenotypic feature that was recently reported in another affected individual with a de novo NR2F2 variant [13]. Interestingly, foregut mesenchyme specific conditional Nr2f2−/− mice were previously shown to have increased perinatal mortality, CDH, and spleen defects including asplenia [10]. Also, vascular malformations, a phenotype seen in the NR2F2 mouse model [4], was seen in two individuals of our cohort (individuals 4 and 13). Nr2f2−/− mice showed defects in angiogenesis and vascular remodeling, including enlarged blood vessels, abnormal development of the atria and sinus venosus, malformed cardinal veins, and decrease in the complexity of microvasculature in the head and spine regions [4].
Differential diagnoses that were considered for these individuals included Noonan syndrome, Prader-Willi syndrome, Pitt-Hopkins syndrome, Fryns syndrome, Smith-Lemli-Opitz syndrome, peroxisomal disorders, Russell-Silver syndrome, geleophysic dysplasia, Moore Federman syndrome, Myhre syndrome, DSD, and laterality defects.
Genotype-phenotype correlation
Review of all known NR2F2 SNVs, did not demonstrate any genotype-phenotype correlation or hotspot region(s) where missense variants were significantly enriched (Fig. 1). However, many missense variants are localized to a narrow region in the DNA binding domain (aa 86–105; Fig. 1). Interestingly, the 46,XX DSD associated NR2F2 LoF variants were all early truncations in coding exon 1 (individual 15, [20]; Fig. 1]) or entire gene deletion [27], whereas, on the contrary, all the affected 46,XX individuals with NR2F2 early truncations [16, 23] or gene deletion (individual 12) did not present with a DSD phenotype (Fig. 1 and Supplementary Table 1).
Our cohort had five predicted LoF NR2F2 SNVs, and they were present in individuals with a syndromic CHD presentation with neurodevelopmental issues (individuals 5-1, 6, 10 and 13), or 46,XX ovotesticular DSD (individual 15). Similar to our cohort, NR2F2 LoF variants have been previously reported in cases with CHD, CDH, and syndromic 46,XX ovotesticular DSD (Supplementary Table 1) [1, 13–25].
In our cohort, there were ten 46,XX individuals, nine of whom were phenotypic females whereas one individual, who carried a heterozygous NR2F2 LoF variant, was assigned male gender at birth and had a diagnosis of ovotesticular DSD (individual 15; Table 1). NR2F2 LoF variants and a NR2F2 gene deletion have been previously reported in individuals with 46,XX ovotesticular DSD [20, 27]. However, individual 12, a 46,XX phenotypic female, who carried a single copy genomic deletion encompassing the entire NR2F2 gene, did not show evidence of genital anomalies or DSD, but presented with CHD, developmental delays, feeding difficulties, and dysmorphic features. A similar case with a NR2F2 gene deletion has been reported [28].
The three known deletion CNVs encompassing NR2F2 also had varied phenotypes, including syndromic CHD (individual 12), syndromic DSD without CHD [27], and syndromic CHD with CDH [28] (Supplementary Fig. 1). These clinical phenotypes highlight the variable expressivity in NR2F2-associated disorders. In previous studies the CDH phenotype has only been associated with de novo NR2F2 LoF SNVs [15, 21, 23, 24] or gene deletion [27], whereas in our cohort, de novo NR2F2 missense variants in the DNA binding domain were seen in the two individuals affected with CDH (individuals 10 and 11).
A comparison of phenotypes between our cases with missense variants in the DNA binding domain (individuals 2,3,7,8,10,11,14) and in the ligand binding domain (individuals 1,9,16) did not yield any genotype phenotype correlation. Larger cohorts may be needed to recognize any association.
Two of the NR2F2 variants in our cohort affect amino acid Cys96 (p.(Cys96Phe), p.(Cys96Tyr)), and a nonsense variant at the same amino acid residue has been reported in a CHD cohort [17]. p.(Cys96Ter) variant was identified in a family with six affected individuals with bicuspid aortic valve and fusion of the right and left coronary aortic valve cusps. Two of the affected individuals had aortic stenosis along with aortic regurgitation and another two affected individuals had VSD in addition to bicuspid aortic valve. The two affected individuals in our cohort (individuals 2 and 8) with de novo variants altering Cys98, had CHD, developmental delays, feeding difficulties, and dysmorphic facial features. Individual 2 also had multicystic dysplastic kidneys with chronic renal failure.
Interestingly, two NR2F2 variants have each been observed recurrently in three unrelated, affected individuals (p.(Pro33AlafsTer77) and p.(Ser341Tyr)). The p.(Pro33AlafsTer77) LoF variant has been reported in two individuals with CHD and 46,XX ovotesticular DSD and in one individual with CHD and CDH [20, 23]. For one of the individuals with 46,XX ovotesticular DSD, the inheritance of p.(Pro33AlafsTer77) was unknown, and for the remaining two cases it was de novo. Similarly, the de novo p.(Ser341Tyr) variant was seen in one individual with an atrioventricular septal defect [1] and in a second individual with asplenia, developmental delays, dysmorphic features, frequent infections, bilateral cryptorchidism, and glandular hypospadias without CHD [13]. In our cohort, this variant was seen in an individual with an atrioventricular defect, feeding difficulties, developmental delays and dysmorphic facial features (individual 9); however, the inheritance of this variant was unknown (not maternal). These variable phenotypic manifestations in individuals carrying identical variants underscore the variable expressivity associated with NR2F2 sequence alterations. Additional genetic and environmental modifiers may have played a role in the phenotypic expression of the NR2F2-related disorder.
Based on the clinical features described in our study, we recommend that any individual with a pathogenic/likely pathogenic NR2F2 variant should have the following evaluations (1) echocardiogram and electrocardiogram (2) renal-bladder ultrasound (3) assessment of external genitalia and consideration of imaging if ambiguous (pelvic ultrasound for females/genital tract ultrasound for males) (4) audiology evaluation (5) ophthalmology examination (6) consideration of early placement of gastric tube for problems with weight gain and oral feeding (7) early intervention for speech, occupational, and physical therapy (8) neuropsychological evaluation and developmental assessment as soon as diagnosis is made and (9) confirmation of sex chromosome complement. Testing for NR2F2 should be considered in any syndromic CHD patient, especially with BPES facial features.
In conclusion, we report findings of 17 unreported individuals and summarize previously reported individuals with heterozygous variants in NR2F2, to provide a comprehensive overview of the spectrum of associated features. Based on these data, we provide clinical recommendations for assessment of affected individuals upon diagnosis. These data and recommendations provide guidance for the clinicians and laboratory personnel who encounter this rare disorder.
Supplementary information
Acknowledgements
We would like to thank all the families who participated in this study. The National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination and the Office of the NIH Director, the Vanderbilt University Medical Center clinical site (U01HG007674).
Author contributions
MG collected, analyzed and interpreted the data, drafted the introduction, results, discussion, figures, and tables. MG, LSM, MM, DL, EB, SBS, SMW, KL, PA, AS, LB, ATW, JMS, KS, FD, BC, BI, MFB, AP, JA, EZ, JPS, ADI, JM, VS, SL, SB, IT, JDC, CTG, WKC, SB, EB, NCB, AS provided the clinical data, wrote the clinical case descriptions, methods, critically reviewed and edited the manuscript. EB conceived the study, interpreted the data, and critically reviewed the manuscript.
Funding
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study was approved by the UK Research Ethics Committee (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. Wendy Chung acknowledges funding support from grants P01HD068250 and U01 HL131003. Sara Sanz Benito acknowledges funding from the Fondo de Investigación Sanitaria (PI15/01647 [SB-S]). Maria Francesca Bedeschi, Nuria C Bramswig and Ariane Schmetz are members of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability ERN-ITHACA [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. Nuria C Bramswig and Ariane Schmetz acknowledge that this work was partly done within the Zentrum für Seltene Erkrankungen of the University Hospital Düsseldorf (ZSED).
Data availability
The data in this study is available in the manuscript, Supplementary information, tables, and figures. The NR2F2 variants reported in this study are submitted in ClinVar. The following variants have been previously submitted by the clinical testing labs/research groups in ClinVar NM_021005.4: c.1019del:p.(Lys340SerfsTer39), c.746G>A:p.(Trp249Ter), c.558dup:p.(Arg187AlafsTer122), c.269A>G:p.(His90Arg), c.1022C>A:p.(Ser341Tyr), c.1097G>C:p.(Arg366Pro) (Variation IDs: 2429770, 598763, 1805610, 1064859, 128232, 521133). The details of the two variants - c.287G>T:p.(Cys96Phe), c.257C>G:p.(Ser86Trp) which were ascertained as part of the DDD study are available in the DECIPHER website (https://www.deciphergenomics.org/patient/259383/genotype/191257/browser; https://www.deciphergenomics.org/patient/282004/genotype/197133/browser). The Clinvar IDs for the remaining variants are listed below (Variation IDs: 2570648, 2570646, 2570643, 2570644, 2570645, 2570642, 2570647).
Competing interests
The authors declare no competing interests.
Ethical approval
The Institutional Review Board of the Children’s Hospital of Philadelphia approved this study. Informed consent was obtained from all individual participants included in the study. Families of individuals 2 &12 consented for publication of images.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Contributor Information
Elizabeth Bhoj, bhoje@email.chop.edu.
Undiagnosed Diseases Network:
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-023-01434-5.
References
- 1.Al Turki S, Manickaraj AK, Mercer CL, Gerety SS, Hitz MP, Lindsay S, et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am J Hum Genet. 2014;94:574–85. doi: 10.1016/j.ajhg.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polvani S, Pepe S, Milani S, Galli A. COUP-TFII in health and disease. Cells. 2019;9:101. doi: 10.3390/cells9010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. Coup d'Etat: an orphan takes control. Endocr Rev. 2011;32:404–21. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–49. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao F, Franco HL, Rodriguez KF, Brown PR, Tsai MJ, Tsai SY, et al. Elimination of the male reproductive tract in the female embryo is promoted by COUP-TFII in mice. Science. 2017;357:717–20. doi: 10.1126/science.aai9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurihara I, Lee DK, Petit FG, Jeong J, Lee K, Lydon JP, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007;3:e102. doi: 10.1371/journal.pgen.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petit FG, Jamin SP, Kurihara I, Behringer RR, DeMayo FJ, Tsai MJ, et al. Deletion of the orphan nuclear receptor COUP-TFII in uterus leads to placental deficiency. Proc Natl Acad Sci USA. 2007;104:6293–8. doi: 10.1073/pnas.0702039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takamoto N, Kurihara I, Lee K, Demayo FJ, Tsai MJ, Tsai SY. Haploinsufficiency of chicken ovalbumin upstream promoter transcription factor II in female reproduction. Mol Endocrinol. 2005;19:2299–308. doi: 10.1210/me.2005-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamoto N, You LR, Moses K, Chiang C, Zimmer WE, Schwartz RJ, et al. COUP-TFII is essential for radial and anteroposterior patterning of the stomach. Development. 2005;132:2179–89. doi: 10.1242/dev.01808. [DOI] [PubMed] [Google Scholar]
- 10.You LR, Takamoto N, Yu CT, Tanaka T, Kodama T, Demayo FJ, et al. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2005;102:16351–6. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arsov T, Kelecic J, Frkovic SH, Sestan M, Kifer N, Andrews D, et al. Expanding the clinical spectrum of pathogenic variation in NR2F2: asplenia. Eur J Med Genet. 2021;64:104347. doi: 10.1016/j.ejmg.2021.104347. [DOI] [PubMed] [Google Scholar]
- 14.Qiao XH, Wang Q, Wang J, Liu XY, Xu YJ, Huang RT, et al. A novel NR2F2 loss-of-function mutation predisposes to congenital heart defect. Eur J Med Genet. 2018;61:197–203. doi: 10.1016/j.ejmg.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Reuter MS, Chaturvedi RR, Liston E, Manshaei R, Aul RB, Bowdin S, et al. The Cardiac Genome Clinic: implementing genome sequencing in pediatric heart disease. Genet Med. 2020;22:1015–24. doi: 10.1038/s41436-020-0757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter F, Morton SU, Kim SW, Kitaygorodsky A, Wasson LK, Chen KM, et al. Genomic analyses implicate noncoding de novo variants in congenital heart disease. Nat Genet. 2020;52:769–77. doi: 10.1038/s41588-020-0652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Abhinav P, Xu YJ, Li RG, Zhang M, Qiu XB, et al. NR2F2 loss‑of‑function mutation is responsible for congenital bicuspid aortic valve. Int J Mol Med. 2019;43:1839–46. doi: 10.3892/ijmm.2019.4087. [DOI] [PubMed] [Google Scholar]
- 18.Upadia J, Gonzales PR, Robin NH. Novel de novo pathogenic variant in the NR2F2 gene in a boy with congenital heart defect and dysmorphic features. Am J Med Genet A. 2018;176:1423–26. doi: 10.1002/ajmg.a.38700. [DOI] [PubMed] [Google Scholar]
- 19.Kammoun M, Souche E, Brady P, Ding J, Cosemans N, Gratacos E, et al. Genetic profile of isolated congenital diaphragmatic hernia revealed by targeted next-generation sequencing. Prenat Diagn. 2018;38:654–63. doi: 10.1002/pd.5327. [DOI] [PubMed] [Google Scholar]
- 20.Bashamboo A, Eozenou C, Jorgensen A, Bignon-Topalovic J, Siffroi JP, Hyon C, et al. Loss of function of the nuclear receptor NR2F2, encoding COUP-TF2, causes testis development and cardiac defects in 46,xx children. Am J Hum Genet. 2018;102:487–93. doi: 10.1016/j.ajhg.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao L, Wynn J, Yu L, Hernan R, Zhou X, Duron V, et al. Likely damaging de novo variants in congenital diaphragmatic hernia patients are associated with worse clinical outcomes. Genet Med. 2020;22:2020–28. doi: 10.1038/s41436-020-0908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li AH, Hanchard NA, Furthner D, Fernbach S, Azamian M, Nicosia A, et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Med. 2017;9:95. doi: 10.1186/s13073-017-0482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.High FA, Bhayani P, Wilson JM, Bult CJ, Donahoe PK, Longoni M. De novo frameshift mutation in COUP-TFII (NR2F2) in human congenital diaphragmatic hernia. Am J Med Genet A. 2016;170:2457–61. doi: 10.1002/ajmg.a.37830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunami N, Shanmugam H, Baird L, Stevens J, Byrne JL, Barnhart DC, et al. Germline but not somatic de novo mutations are common in human congenital diaphragmatic hernia. Birth Defects Res. 2018;110:610–17. doi: 10.1002/bdr2.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab ME, Dong S, Lianoglou BR, Aguilar Lucero AF, Schwartz GB, Norton ME, et al. Exome sequencing of fetuses with congenital diaphragmatic hernia supports a causal role for NR2F2, PTPN11, and WT1 variants. Am J Surg. 2022;223:182–86. doi: 10.1016/j.amjsurg.2021.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Zidoune H, Ladjouze A, Chellat-Rezgoune D, Boukri A, Dib SA, Nouri N, et al. Novel genomic variants, atypical phenotypes and evidence of a digenic/oligogenic contribution to disorders/differences of sex development in a large North African cohort. Front Genet. 2022;13:900574. doi: 10.3389/fgene.2022.900574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalheira G, Malinverni AM, Moyses-Oliveira M, Ueta R, Cardili L, Monteagudo P, et al. The natural history of a man with ovotesticular 46,XX DSD caused by a novel 3-Mb 15q26.2 deletion containing NR2F2 gene. J Endocr Soc. 2019;3:2107–13. doi: 10.1210/js.2019-00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady PD, DeKoninck P, Fryns JP, Devriendt K, Deprest JA, Vermeesch JR. Identification of dosage-sensitive genes in fetuses referred with severe isolated congenital diaphragmatic hernia. Prenat Diagn. 2013;33:1283–92. doi: 10.1002/pd.4244. [DOI] [PubMed] [Google Scholar]
- 29.Poot M, Verrijn Stuart AA, van Daalen E, van Iperen A, van Binsbergen E, Hochstenbach R. Variable behavioural phenotypes of patients with monosomies of 15q26 and a review of 16 cases. Eur J Med Genet. 2013;56:346–50. doi: 10.1016/j.ejmg.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Mosca AL, Pinson L, Andrieux J, Copin H, Bigi N, Puechberty J, et al. Refining the critical region for congenital diaphragmatic hernia on chromosome 15q26 from the study of four fetuses. Prenat Diagn. 2011;31:912–4. doi: 10.1002/pd.2793. [DOI] [PubMed] [Google Scholar]
- 31.Dateki S, Fukami M, Tanaka Y, Sasaki G, Moriuchi H, Ogata T. Identification of chromosome 15q26 terminal deletion with telomere sequences and its bearing on genotype-phenotype analysis. Endocr J. 2011;58:155–9. doi: 10.1507/endocrj.K10E-251. [DOI] [PubMed] [Google Scholar]
- 32.Rump P, Dijkhuizen T, Sikkema-Raddatz B, Lemmink HH, Vos YJ, Verheij JB, et al. Drayer’s syndrome of mental retardation, microcephaly, short stature and absent phalanges is caused by a recurrent deletion of chromosome 15(q26.2—>qter) Clin Genet. 2008;74:455–62. doi: 10.1111/j.1399-0004.2008.01064.x. [DOI] [PubMed] [Google Scholar]
- 33.Davidsson J, Collin A, Bjorkhem G, Soller M. Array based characterization of a terminal deletion involving chromosome subband 15q26.2: an emerging syndrome associated with growth retardation, cardiac defects and developmental delay. BMC Med Genet. 2008;9:2. doi: 10.1186/1471-2350-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poot M, Eleveld MJ, van 't Slot R, van Genderen MM, Verrijn Stuart AA, Hochstenbach R, et al. Proportional growth failure and oculocutaneous albinism in a girl with a 6.87 Mb deletion of region 15q26.2—>qter. Eur J Med Genet. 2007;50:432–40. doi: 10.1016/j.ejmg.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Rujirabanjerd S, Suwannarat W, Sripo T, Dissaneevate P, Permsirivanich W, Limprasert P. De novo subtelomeric deletion of 15q associated with satellite translocation in a child with developmental delay and severe growth retardation. Am J Med Genet A. 2007;143A:271–6. doi: 10.1002/ajmg.a.31581. [DOI] [PubMed] [Google Scholar]
- 36.Hoang TT, Goldmuntz E, Roberts AE, Chung WK, Kline JK, Deanfield JE, et al. The congenital heart disease genetic network study: cohort description. PLoS One. 2018;13:e0191319. doi: 10.1371/journal.pone.0191319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pediatric Cardiac Genomics C, Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, et al. The congenital heart disease genetic network study: rationale, design, and early results. Circ Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rastetter RH, Bernard P, Palmer JS, Chassot AA, Chen H, Western PS, et al. Marker genes identify three somatic cell types in the fetal mouse ovary. Dev Biol. 2014;394:242–52. doi: 10.1016/j.ydbio.2014.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in this study is available in the manuscript, Supplementary information, tables, and figures. The NR2F2 variants reported in this study are submitted in ClinVar. The following variants have been previously submitted by the clinical testing labs/research groups in ClinVar NM_021005.4: c.1019del:p.(Lys340SerfsTer39), c.746G>A:p.(Trp249Ter), c.558dup:p.(Arg187AlafsTer122), c.269A>G:p.(His90Arg), c.1022C>A:p.(Ser341Tyr), c.1097G>C:p.(Arg366Pro) (Variation IDs: 2429770, 598763, 1805610, 1064859, 128232, 521133). The details of the two variants - c.287G>T:p.(Cys96Phe), c.257C>G:p.(Ser86Trp) which were ascertained as part of the DDD study are available in the DECIPHER website (https://www.deciphergenomics.org/patient/259383/genotype/191257/browser; https://www.deciphergenomics.org/patient/282004/genotype/197133/browser). The Clinvar IDs for the remaining variants are listed below (Variation IDs: 2570648, 2570646, 2570643, 2570644, 2570645, 2570642, 2570647).