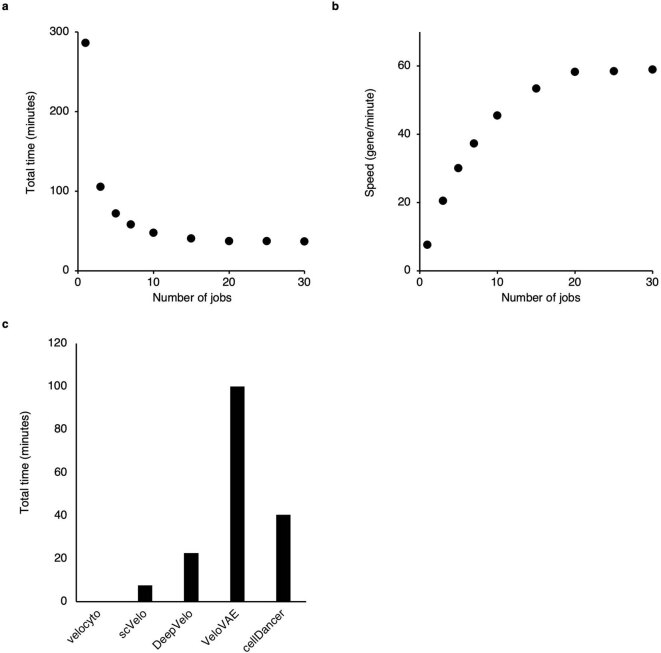
Extended Data Fig. 10. The speedup of cellDancer.
(a, b) Scatter plots showing the total time (a) and the training speed (b) of cellDancer when applying multiprocessing. We tested the parallel speedup ratio of cellDancer by increasing job numbers from 1 to 30. We applied full analysis of cellDancer to 2,159 genes in 18,140 cells (the hippocampal dentate gyrus neurogenesis dataset) with the default parameters and calculated the runtime and speed of different job numbers. The evaluation of all the algorithms and the speedup ratio analysis was performed on a 2.7 GHz 24-Core Intel Xeon W processor. Total runtime decreases from 286 to 36 minutes when adding job numbers from 1 to 30 and reaches saturation at 15 jobs with 40 minutes. cellDancer has a feasible runtime of 53 genes per minute using 15 jobs. The training speed (number of genes per unit time) increases with the number of jobs. (c) Bar plot showing the total time of the comparison between velocyto, scVelo, DeepVelo, VeloVAE, and cellDancer. We compared the runtime of cellDancer with velocyto, scVelo, DeepVelo, and VeloVAE. The benchmark is based on 18,140 cells and 2,159 genes in the hippocampal dentate gyrus neurogenesis dataset with the default parameters. We set the number of jobs (threads) to 15 for scVelo, DeepVelo, VeloVAE, and cellDancer. cellDancer shows a comparable running time with the other two deep learning algorithms.