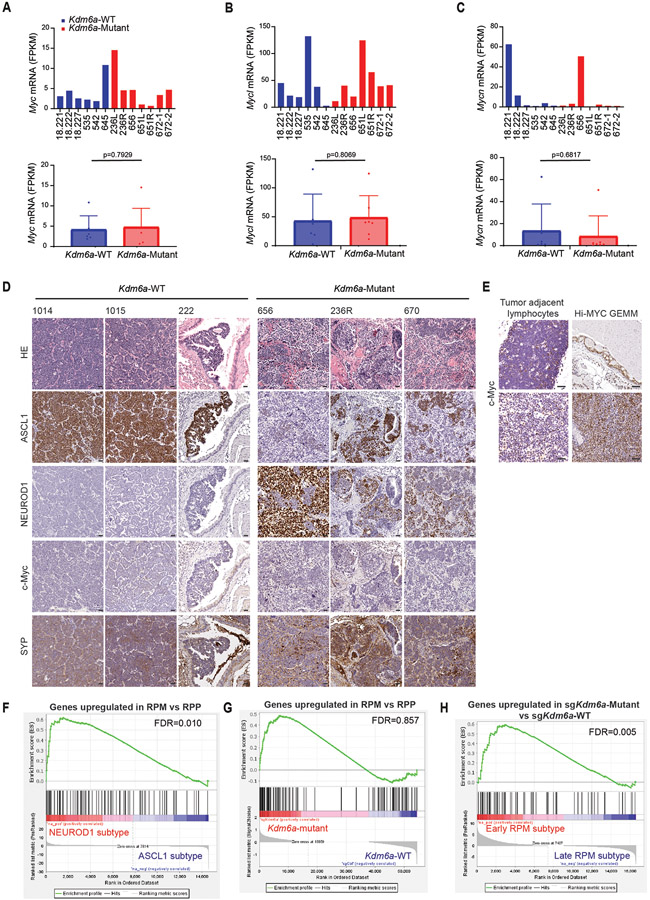
Extended Data Fig. 5. Comparison of Kdm6a-Mutant SCLC GEMM with the Myc-driven RPM SCLC GEMM using RNA-seq and IHC from tumors.
(a-c) mRNA expression from the RNA-seq data (from Fig. 1d) for Myc (a), Mycl (b) and Mycn (c) of individual Kdm6a-WT and Kdm6a-Mutant mouse SCLC tumors (top) or combined tumors by genotype (bottom). See Supplementary Table 2. For a,b,c, data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated on each figure panel. n=6 Kdm6a-WT tumors from independent mice and n=7 Kdm6a-Mutant tumors. (d) H&E and IHC staining for ASCL1, NEUROD1, c-Myc and Synaptophysin from 3 Kdm6a-WT (1014, 1015, 222) and 3 Kdm6a-Mutant (656, 236R, 670) mouse SCLC lung tumors. Scale Bar=50 μm. (e) IHC staining for c-Myc from tumor adjacent lymphocytes and high c-Myc prostate cancer GEMM as positive control of c-Myc staining. Scale Bar=50 μm. (f-h) GSEA of genes upregulated in the RPM vs. RPP GEMM13 with publicly available scRNA-seq data from human SCLCs of the ASCL1 and NEUROD1 subtypes34 (see Methods) (f), or with RNA-seq data from Kdm6a-Mutant vs. Kdm6a-WT tumors from Fig. 1d (g), or using GSEA of genes upregulated in Kdm6a-Mutant tumors (see Fig. 1e) with late RPM vs. early RPM subtypes13. FDR q-values from GSEA adjusted for multiple hypothesis testing are indicated.