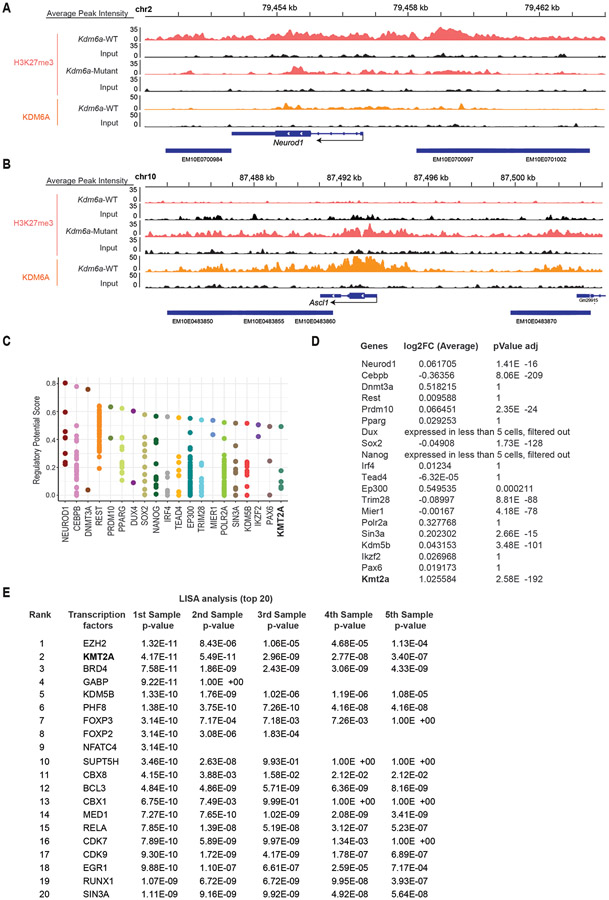
Extended Data Fig. 9. NEUROD1 Expression after KDM6A Inactivation is Partially Mediated by KMT2A.
(a,b) Tracks of H3K27me3 and KDM6A ChIP-seq of Neurod1 (a) and Ascl1 (b) from ChIP-seq data from Fig. 5. Each track is the sum of the peaks of: 2 Kdm6a-WT tumor-derived cell lines for Kdm6a-WT (1014, 159-1) and 2 Kdm6a-Mutant tumor-derived cell lines for Kdm6a-Mutant (236L, 236R) with their respective input. (c) Cistrome analysis of transcription factors and chromatin regulators of Neurod1 gene in human hg38 (http://dbtoolkit.cistrome.org/) within 10 kilobases of the Neurod1 gene. (d) Pseudo-bulk differentially expressed gene (DEG) analysis from the scRNA-seq data in Fig. 3i in Kdm6a-Mutant vs Kdm6a-WT tumors cells of the genes in c. Non-parametric Wilcoxon rank sum test was used and p-values adjusted for multiple hypothesis testing are shown. (e) List of the top 20 candidate regulators of accessible peaks determined by LISA analysis of the 100 top differentially accessible peaks at TSSs with LFC>1 sorted by ascending p-value in Kdm6a-Mutant vs. Kdm6a-WT using ATAC-seq data from Fig. 2a (see Supplementary Table 8, tab 1 for complete list).