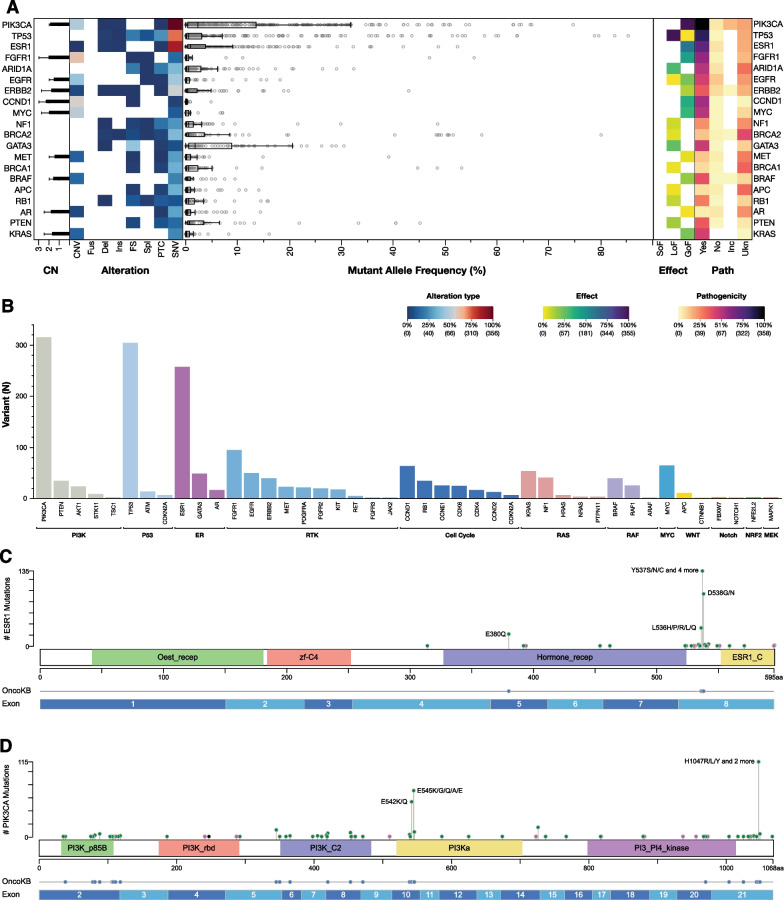
Fig. 1.
Landscape plot of all detectable aberrations in ctDNA samples. A Incidence of the single aberrations [copy number variations (CNV), Fusions (Fus), deletions (Del), insertions (Ins), frameshift (FS), splicing variants (Spl), premature termination codons (PTC) and single nucleotide variation (SNV)] is represented on the left. The mutant allele frequency (MAF) of each mutation is shown in the middle. Effect [gain of function (GOF), loss of function (LOF) and switch of function (SOF)] and pathogenicity [yes, no, unknown (Ukn) and inconclusive (Inc)] of all the detected aberrations are show on the right. Histogram representing different frequency distribution of gene mutations across oncogenic pathways (B). Lollipop plot showing the distribution of ESR1 (C) and PIK3CA (D) mutations by their amino acid coordinates across ER and PI3K domains and across ESR1 and PIK3CA exon sequencies. Oncogenic mutations are highlighted