In this study, Qian et al. generated a novel monoclonal antibody, RD-43, targeting the ectodomain of protein tyrosine phosphatase receptor δ (PTPRD) to promote PTPRD dimerization, which inhibits PTPRD function and promotes its degradation. They show that treatment with RD-43 suppresses SRC signaling required for breast cancer cell invasion, and their findings demonstrate that antibodies targeted to the extracellular segments of receptor protein tyrosine phosphatases may have clinical applications.
Keywords: bivalent antibody, dimerization, PTPRD, protein tyrosine phosphorylation, receptor protein tyrosine phosphatase, SRC
Abstract
Protein tyrosine phosphatases (PTPs) are critical regulators of signal transduction but have yet to be exploited fully for drug development. Receptor protein tyrosine phosphatase δ (RPTPδ/PTPRD) has been shown to elicit tumor-promoting functions, including elevating SRC activity and promoting metastasis in certain cell contexts. Dimerization has been implicated in the inhibition of receptor protein tyrosine phosphatases (RPTPs). We have generated antibodies targeting PTPRD ectodomains with the goal of manipulating their dimerization status ectopically, thereby regulating intracellular signaling. We have validated antibody binding to endogenous PTPRD in a metastatic breast cancer cell line, CAL51, and demonstrated that a monoclonal antibody, RD-43, inhibited phosphatase activity and induced the degradation of PTPRD. Similar effects were observed following chemically induced dimerization of its phosphatase domain. Mechanistically, RD-43 triggered the formation of PTPRD dimers in which the phosphatase activity was impaired. Subsequently, the mAb–PTPRD dimer complex was degraded through lysosomal and proteasomal pathways, independently of secretase cleavage. Consequently, treatment with RD-43 inhibited SRC signaling and suppressed PTPRD-dependent cell invasion. Together, these findings demonstrate that manipulating RPTP function via antibodies to the extracellular segments has therapeutic potential.
Protein tyrosine phosphorylation is a reversible, post-translational modification orchestrated by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs). Owing to the wide variety of cellular events regulated by modulation of tyrosine phosphorylation status, both PTKs and PTPs have long been considered attractive pharmacological targets (Krebs 1993). The development of PTK inhibitors has yielded numerous successful drugs, highlighting the promise of exploiting dysfunctional tyrosine phosphorylation-dependent signal transduction for therapeutic development in various diseases, including cancer. In contrast, whereas PTPs also play indispensable roles in balancing cellular signaling pathways and have been implicated in multiple diseases (Tonks 2013), they have been historically underexploited as therapeutic targets, mostly due to their highly charged catalytic pocket. Small molecule inhibitors targeting the PTP active site are predominantly charged molecules, which underlies poor bioavailability (Mullard 2018). Hence, alternative approaches, such as targeting allosteric sites (Krishnan et al. 2014; Chen et al. 2016), stabilizing the inactive oxidized form of PTPs (Haque et al. 2011), or developing cell-permeable peptides (Lang et al. 2015), are being pursued.
Receptor protein tyrosine phosphatases (RPTPs) are transmembrane proteins comprising an extracellular receptor segment and an intracellular phosphatase segment. RPTPs have two intracellular phosphatase domains: The membrane-proximal domain (D1) performs the catalytic function, whereas the membrane-distal domain (D2) has no catalytic function but regulates RPTP activity (Tonks 2006). Receptor PTKs are activated by dimerization, and antibodies that perturb formation of RTK dimers inhibit the kinase activity (Fauvel and Yasri 2014). In a complementary manner, dimerization has been shown to inhibit RPTP activity (van der Wijk et al. 2005). Therefore, pharmacological agents engaging the extracellular segment of RPTPs offer an innovative approach to manipulate the function of these receptor proteins.
Diverse mechanisms have been proposed to explain dimerization-mediated inhibition of RPTPs, involving the wedge motif (Bilwes et al. 1996; Majeti et al. 1998), D2 domain (Fujikawa et al. 2019; Wen et al. 2020), and transmembrane domain (Chin et al. 2005; Bloch et al. 2019). The wedge motif is a conserved helix–turn–helix located between the transmembrane region and the D1 domain in some members of the RPTP family. The wedge motif has been shown to interact with the catalytic pocket of the apposing D1 domain in crystal structures (Bilwes et al. 1996), and mutations at this interface have been shown to impair dimerization-associated inhibition in RPTPs (Majeti et al. 1998). These models provide a structural basis for the regulation of RPTP function by intrinsic homophilic interaction (Tertoolen et al. 2001; Xu and Weiss 2002) or ligand-mediated monomer/dimer/oligomer switches (Fukada et al. 2006; Coles et al. 2011) under physiological conditions, indicating that dimerization-mediated inhibition may be a shared regulatory feature across the RPTP family. Consequently, targeting of the ectodomain is attractive as the basis for modulating RPTP function pharmacologically. Nevertheless, there are several critical biochemical details that remain to be characterized, such as the fate of dimerized RPTP molecules and whether secretase cleavage, which is shown to modify the extracellular segments of RPTPs and result in shedding, may affect the efficacy of approaches to induce artificial dimerization via the extracellular segments.
Receptor protein tyrosine phosphatase δ (PTPRD) is primarily expressed in the central nervous system (Cornejo et al. 2021); however, aberrant expression of PTPRD is observed in a subset of metastatic breast cancer patients, particularly in those deficient in expression of metastasis suppressor protein 1 (MTSS1), a putative tumor suppressor gene (Chaudhary et al. 2015) and a binding partner of PTPRD (Woodings et al. 2003). Several studies suggest that PTPRD is associated with invasive morphological changes (Chaudhary et al. 2015) and hyperactivation of the proto-oncogenic protein tyrosine kinase SRC (Yeatman 2004), consistent with an oncogenic role in certain contexts. For example, PTPRD overexpression promotes invasive growth of MCF10A cells in vitro (Chaudhary et al. 2015). Similarly, in xenografts, PTPRD levels are positively correlated with the metastasis of breast cancer cells (Yuwanita et al. 2015). In mouse models, PTPRD depletion attenuates the activity of SRC family kinases (Nakamura et al. 2017) and perturbs the self-renewal of neural progenitor cells (Tomita et al. 2020). Mechanistically, this results from PTPRD dephosphorylation of phospho-tyrosine 527 (pTyr527) located in the C-terminal autoinhibiting loop of SRC. Dephosphorylation of pTyr527 triggers autophosphorylation of tyrosine 416 (pTyr416) within the SRC kinase domain, which ultimately stimulates SRC activity (Hunter 2015). Therefore, inhibition of PTPRD alone or in combination with anti-SRC therapy may offer a path for intervention in a subset of metastatic breast cancers.
PTPRD, together with PTPRS and PTPRF (also known as LAR), belongs to the LAR-RPTP subfamily (Cornejo et al. 2021). The extracellular segment of LAR-like RPTPs is composed of three immunoglobulin-like domains (Ig) and four or eight fibronectin III-like domains (FNIII), depending on the isoform (Pulido et al. 1995). In addition, there are three secretase cleavage sites that modify the extracellular and transmembrane domains (Fig. 1A). Secretase cleavage serves critical roles in the translocation and destruction of RPTPs. In the secretory pathway, the ectodomain is proteolytically processed by a furin family secretase (S1) and forms a complex through noncovalent interactions between the resulting segments (Fig. 1A; Anders et al. 2006). Two additional cleavage events, S2 and S3, regulate the subcellular localization and stability of LAR-like RPTPs (Cornejo et al. 2021) through a mechanism similar to that of other membrane proteins, such as Notch (Shih and Wang 2007) and PTKs (Merilahti and Elenius 2019). S2 cleavage, also known as shedding, releases the ectodomain of receptor phosphatases from the cell membrane and promotes S3 cleavage, as well as the degradation of RPTP molecules (Aicher et al. 1997; Burden-Gulley et al. 2010). The S3 cleavage event releases the phosphatase intracellular domain (PICD) into the cytosol, where it remains catalytically active but is subject to facilitated degradation (Haapasalo et al. 2007; Phillips-Mason et al. 2011). It remains to be determined whether secretase-dependent degradation is the only mechanism for RPTP destruction from the membrane and whether artificial dimerization of the ectodomains may affect the intracellular catalytic domains without being compromised by these cleavage events.
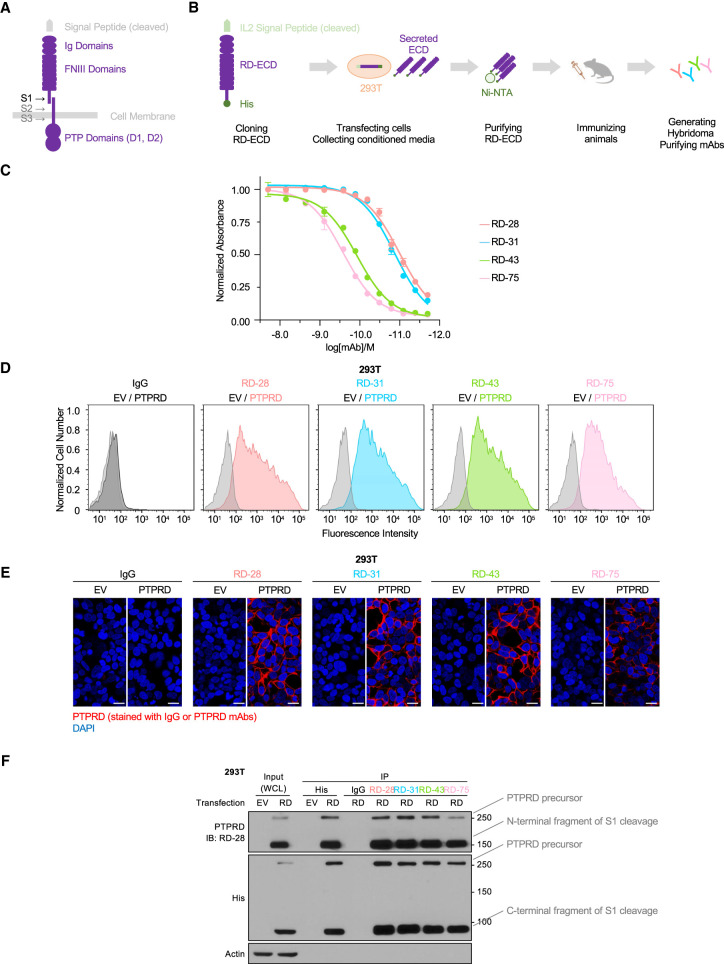
Figure 1.
Monoclonal antibodies targeting PTPRD ectodomain were generated and validated. (A) Schematic illustration of PTPRD (isoform 1) structure. (Ig domains) Immunoglobulin-like domains, (FNIII domains) fibronectin type III-like domains, (PTP domains) protein tyrosine phosphatase domains. S1, S2, and S3 represent sites of proteolytic processing. (B) Schematic illustration of the workflow to produce PTPRD ectodomain (RD-ECD) as antigen to generate antibodies from immunized rats. (C) ELISA measurements of the affinity of PTPRD mAbs for the PTPRD ectodomain. Data are normalized to the highest antibody concentration set at 100%. (D) Flow cytometry analysis of 293T cells transfected with empty vector (EV) or PTPRD stained with IgG or the indicated PTPRD antibodies. (E) Representative confocal immunofluorescence microscopy images of 293T cells stably expressing empty vector (EV) or PTPRD stained with IgG or the indicated PTPRD antibodies (red). Scale bars, 20 μm. (F) Immunoblot analysis of lysates ([WCL] whole-cell lysate) of 293T cells transfected with empty vector (EV) or PTPRD, followed by immunoprecipitates (IP) with anti-His antibody-conjugated beads, or Protein G beads conjugated with IgG or the indicated PTPRD antibodies.
In this study, we have implemented a monoclonal antibody-based strategy to induce PTPRD dimerization ectopically and ultimately suppress PTPRD-dependent cell invasion. We produced and evaluated an array of monoclonal antibodies that target the ectodomain of PTPRD. We validated two cell models in which PTPRD activity was assessed by SRC phosphorylation and regulated by chemically induced dimerization. After demonstrating that antibody RD-43 was capable of inhibiting PTPRD activity and promoting the degradation of PTPRD, we established that RD-43 both induced PTPRD dimerization and facilitated the degradation of PTPRD dimers in a secretase-independent pathway. In a breast cancer cell model, we demonstrated that RD-43 suppressed PTPRD-dependent cell invasion. Collectively, this research highlights the potential of antibodies to the RPTP ectodomains as probes to target their enzymatic functions and modify their stability.
Results
Monoclonal antibodies targeting PTPRD ectodomain were generated and validated
Our approach to manipulate PTPRD activity using antibodies relied on recognition of their ectodomains displayed on the cell surface. To generate PTPRD ectodomain-specific monoclonal antibodies (mAbs), we used recombinant human PTPRD ectodomain secreted from HEK293T cells (293T) as an antigen to generate a library of hybridoma clones (Fig. 1B). Positive mAb clones were screened through enzyme-linked immunoassay (ELISA) (Fig. 1C; Supplemental Fig. S1A). Subdomains of PTPRD-ECD were cloned, expressed, and purified by the same method, and epitopes recognized by the mAbs were characterized by ELISA. Our epitope-mapping data showed that antibody RD-43 recognized the FN domain segment of PTPRD (Supplemental Fig. S1B).
We used various methods to validate the ability of mAbs to recognize cells expressing PTPRD. By flow cytometry, we observed that PTPRD mAbs, but not control IgG, specifically labeled 293T cells expressing PTPRD (Fig. 1D). We used immunofluorescence to validate binding of PTPRD mAbs at the expected membrane locations in fixed 293T cells (Fig. 1E). Moreover, the PTPRD mAb signal colocalized with a commercial anti-His antibody, which targeted the tagged intracellular fragment (Supplemental Fig. S2A) following cell permeabilization. As a complementary approach to assess PTPRD expression by immunoblot, we validated monoclonal antibody RD-28, which detected PTPRD in the denatured form following SDS-PAGE (Supplemental Fig. S2B) and did not crossreact to other LAR-like RPTP proteins (Supplemental Fig. S2C).
Previous studies have indicated that LAR-like RPTPs are proteolytically processed by furin family secretases to generate two noncovalently associated fragments (Fig. 1A; Supplemental Fig. S1; Anders et al. 2006). To characterize this interaction, we performed immunoprecipitation followed by immunoblot with the N-terminal fragment of PTPRD and the His-tagged intracellular fragment. These fragments coimmunoprecipitated in a reciprocal manner (Fig. 1F). Taken together, we generated an array of mAbs that targeted PTPRD with high affinity. We demonstrated that PTPRD is present on the cell surface as a noncovalently associated complex, supporting the structural basis for regulation of PTPRD activity through manipulating its ectodomain. Moreover, we validated the binding of mAbs to PTPRD on the cell membrane by immunofluorescence and flow cytometry and additionally established mAb RD-28 as a probe to assess PTPRD levels by immunoblotting.
PTPRD-dependent SRC activation was reversed by chemically induced dimerization
To explore the modulation of PTPRD function by antibodies, we verified two cell models in which we could assess activity and test whether dimerization inhibited PTPRD. It has been reported previously that PTPRD dephosphorylates the inhibitory pTyr527 of SRC in vitro, which promotes autophosphorylation and activation of the kinase (Chaudhary et al. 2015). Hence, we asked whether PTPRD increased the level of SRC pTyr416, the active form of SRC, in living cells. In transfected 293T cells, we showed a dose-dependent increase of SRC Tyr416 phosphorylation when PTPRD was overexpressed (Fig. 2A). Furthermore, in a metastatic breast cancer cell line, CAL51, CRISPR–CAS9-mediated knockout of PTPRD (sgRD1 and sgRD2) attenuated SRC Tyr416 phosphorylation compared with the scrambled sgRNA control (SCB) (Fig. 2B,C). These cells were chosen because analysis of DepMap RNA-seq data revealed low MTSS1 and high PTPRD levels (Supplemental Fig. S3), consistent with a target tumor type that would be expected to respond to PTPRD antibodies. Similar results were observed by measuring the activation status of SRC with antibodies directed against the nonphosphorylated form of Tyr527 (Fig. 2B,C).
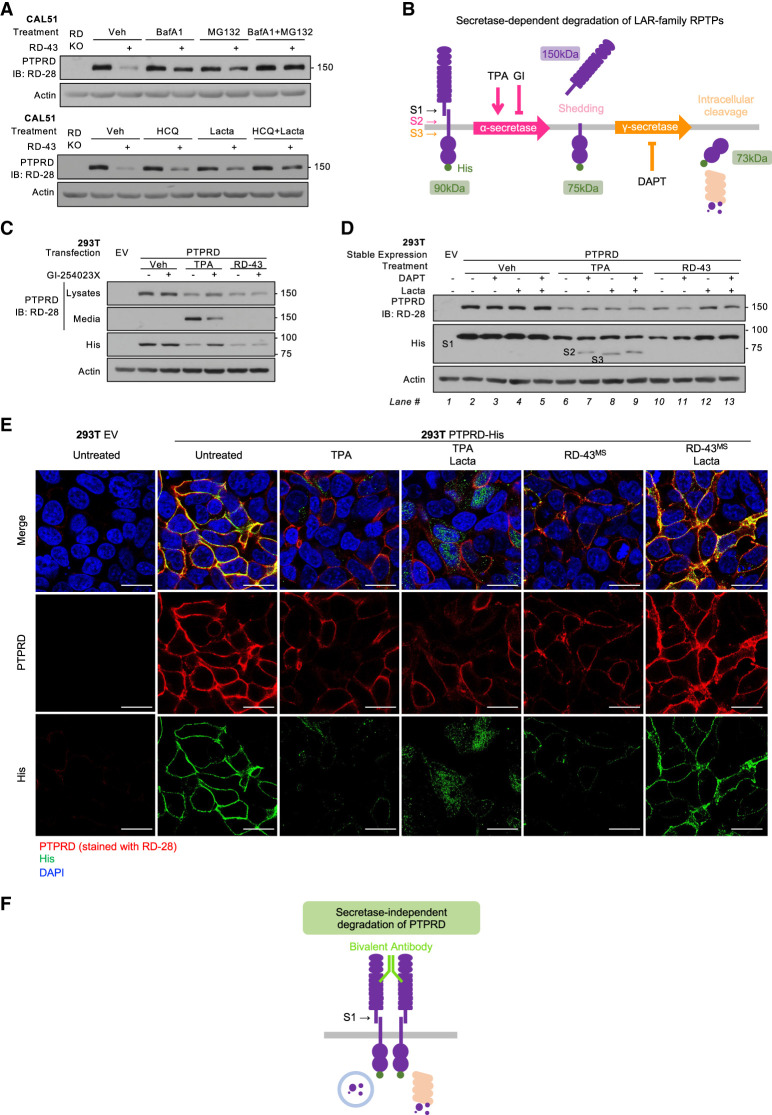
Figure 2.
Chemically induced dimerization inhibited PTPRD-dependent SRC activation. (A) Immunoblot analysis of 293T cells transfected with GFP, PTPRD isoform 1 (full-length PTPRD with eight FNIII domains), or PTPRD isoform 6 (shorter PTPRD with four FNIII domains) at different doses ([+] 1 μg, [++] 2 μg, [+++] 4 μg). (B) Immunoblot analysis of CAL51-CAS9 cells expressing scrambled sgRNA (SCB) or PTPRD targeting sgRNAs sgRD1 and sgRD2. (C) Quantification of densitometry of SRC pTyr416/SRC and SRC non-pTyr527/SRC from three replicates of immunoblot conducted as in B. (***) P-value < 0.001. (D) Schematic illustration of the structure of PTPRD with DmrB and C-terminal His tag. DmrB tag induces homodimer formation in the presence of AP20187. (E) Immunoblot analysis of 293T cells transfected with GFP, DmrB-tagged PTPRD, or catalytically dead DmrB-tagged PTPRD (PTPRD-C/S-DmrB) treated with AP20187 at different doses ([+] 1 μM, [++] 2 μM). (F) Immunoblot analysis of CAL51-CAS9 cells expressing scrambled sgRNA (SCB), PTPRD targeting sgRNA (sgRD-2), and gRNA-resistant PTPRD with DmrB tag (RD*DmrB) treated with AP20187 (2 μM, overnight) or dasatinib (50 nM, 1 h). (G) Quantification of densitometry of SRC pTyr416/SRC from three replicates of immunoblot conducted as in F. (***) P-value < 0.001, (n/s) P-value > 0.05.
To investigate the effect of PTPRD dimerization on SRC activity, we made use of an inducible dimerization system based on the DmrB tag and its synthetic, membrane-permeable ligand, AP20187, which induces the formation of homodimers of DmrB-tagged proteins (Fig. 2D; Yang et al. 2000). In 293T cells, expression of the DmrB-tagged PTPRD activated SRC pTyr416, suggesting that the DmrB tag itself did not interfere with PTPRD activity. Furthermore, in the presence of AP20187, SRC activity was unchanged in cells expressing wild-type, untagged PTPRD; however, we observed an AP20187-dependent decrease in SRC activity in PTPRD-DmrB-expressing cells. In addition, we observed in both mAb RD-28 and anti-His blots that AP20187 treatment resulted in decreased levels of PTPRD protein, indicating that DmrB-mediated dimerization of PTPRD was associated with degradation (Fig. 2E). DmrB-tagged PTPRD carrying a catalytically dead mutation (C1553S [Jia et al. 1995], PTPRD-CS-DmrB) did not activate SRC, indicating that SRC activation was dependent on the catalytic activity of PTPRD. Furthermore, the DmrB-PTPRD-C/S construct was also subject to AP20187-mediated degradation, illustrating that dimerization-associated degradation was independent of its phosphatase activity (Fig. 2E). Moreover, we reconstituted expression of PTPRD using the DmrB-tagged construct (PTPRD*DmrB) in CAL51 PTPRD KO cells and restored SRC pTyr416 phosphorylation to a similar extent compared with the control cells. Again, this effect was abolished by dimerization of PTPRD (Fig. 2F,G), which also promoted the degradation of DmrB-tagged PTPRD in CAL51 cells (Fig. 2F).
Finally, to investigate whether degradation was caused by PTPRD dimerization or SRC inhibition, we treated cells with dasatinib, a small molecule inhibitor of SRC family kinases (Kennedy and Gadi 2018), and measured the levels of PTPRD protein. We showed that dasatinib reduced SRC phosphorylation but did not interfere with PTPRD expression, suggesting that it was dimerization, rather than SRC inhibition, that potentiated PTPRD destruction (Fig. 2F). To summarize, we established a chemically induced system that promoted dimerization of the catalytic fragment of PTPRD. PTPRD-dependent SRC activation was abolished upon induction of dimerization in both 293T cells, in which PTPRD was overexpressed, and CAL51 cells, in which PTPRD was expressed at endogenous levels.
PTPRD mAbs inhibited SRC activity and promoted PTPRD degradation
It is known that the intrinsic bivalency of antibody molecules is capable of inducing protein dimerization (Fauvel and Yasri 2014; Lu et al. 2020). Therefore, we asked whether PTPRD mAbs could recapitulate the SRC inhibition and PTPRD degradation that we observed following chemically induced dimerization.
We performed immunofluorescence staining of fixed CAL51 cells to validate the recognition of endogenous PTPRD by our mAbs. We confirmed that PTPRD mAbs stained wild-type CAL51 cells (RD WT), but not PTPRD knockout cells (RD KO) (Fig. 3A, top panel, fixed staining). Next, we applied these mAbs to live cells, followed by fixation and secondary antibody staining. Interestingly, we observed a loss of signal with mAb RD-43 (Fig. 3A, bottom panel, live staining), indicating that antibody binding may promote PTPRD destruction in living cells. To reveal the dynamics of these effects of RD-43, we performed immunoblot analysis (Fig. 3B). We observed that RD-43 induced noticeable reduction of PTPRD levels following 1-h treatment and further promoted the destruction of PTPRD protein following extended treatment with antibody. Furthermore, treatment with antibody was shown to suppress SRC activity rapidly prior to degradation, indicating that RD-43-bound PTPRD molecules were catalytically impaired before degradation (Fig. 3B). Similar observations were made in 293T cells, where RD-43 treatment reversed PTPRD-dependent signaling changes to the same extent as the DmrB system (Supplemental Fig. S4).
Figure 3.
A bivalent antibody RD-43 inhibited SRC activity and promoted PTPRD degradation. (A) Representative confocal immunofluorescence microscopy images and quantification of CAL51 cells expressing PTPRD (RD WT) or knockout PTPRD (RD KO) stained with representative PTPRD mAbs after fixation (top panel, fixed staining) or incubated with the indicated PTPRD antibodies before fixation (bottom panel, live staining). Relative fluorescence intensities were quantified and normalized to RD-28 staining under either fixed- or live-staining conditions (n = 4). Scale bars, 20 μm. (B) Immunoblot analysis of CAL51 cells that were untreated (CTRL) or treated with 100 nM RD-43 for the indicated duration. (O/N) Overnight. (C) Schematic illustration of the structure of native RD-43, RD-43MS with the mouse-derived Fc region, RD-43MS-Fab with the mouse-derived Fab constant region, and a speculated binding profile of two RD-43MS-Fab molecues with one anti-mouse Fab-specific secondary antibody (anti-Fab 2nd). (D) Immunoblot analysis of CAL51 cells treated with 100 nM rat IgG, RD-43, mouse IgG, or RD-43MS for the indicated durations. (E) Quantification of densitometry of SRC pTyr416/SRC calculated from three replicates of immunoblots conducted as in D. (***) P-value < 0.001, (n/s) P-value > 0.05. (F) Immunoblot analysis of 293T cells transfected with GFP or PTPRD and treated with 100 nM RD-43MS, 200 nM RD-43Fab, or 100 nM anti-mouse Fab-specific secondary antibody (anti-Fab 2nd).
To validate further these effects of RD-43 on SRC inhibition and PTPRD degradation, we tested a murinized isotype of the antibody, in which the native, rat-derived Fc region was swapped with the Fc region from mice (RD-43MS) (Fig. 3C). This murinized isotype enabled us to detect PTPRD simultaneously using both rat isotype RD-28 and RD-43MS antibodies. Under short-term treatment, RD-43MS inhibited SRC activity to the same extent as the native antibody; in long-term treatment, RD-43MS also promoted the loss of PTPRD protein (Fig. 3D).
To test the importance of bivalency in the action of mAb RD-43, we generated the Fab fragment from RD-43MS, which contained only one antigen-binding arm (Fig. 3C). RD-43MS-Fab bound to PTPRD comparably with RD-43MS in ELISA (Supplemental Fig. S5A) and immunofluorescence staining (Supplemental Fig. S5B). However, treatment with RD-43MS-Fab did not cause protein degradation or SRC inhibition in PTPRD-expressing 293T cells (Fig. 3F). Moreover, concomitant treatment of RD-43MS-Fab with an anti-mouse Fab-specific secondary antibody (anti-Fab 2nd), which would be expected to cross-link two Fab molecues, was demonstrated to rescue partially the regulatory functions of RD-43MS-Fab (Fig. 3F; Supplemental Fig. S5B).
Altogether, we demonstrated that a monoclonal antibody, RD-43, as well as its murinized variant, inhibited SRC activity and promoted PTPRD degradation in a manner that was dependent on antibody bivalency, consistent with the results of DmrB-mediated dimerization.
RD-43-bound PTPRD was degraded through lysosomal and proteasomal pathways but independently of secretase cleavage
We have shown that both chemically induced dimerization and RD-43 treatment promoted degradation of PTPRD. To characterize the underlying mechanism, we examined the lysosomal and proteasomal pathways, which coordinate the degradation of membrane proteins (Korolchuk et al. 2010). In CAL51 cells, lysosomal inhibitors bafilomycin A1 (BafA1) (Yamamoto et al. 1998) and hydroxychloroquine (HCQ) (Mauthe et al. 2018) partially rescued RD-43-mediated degradation. Furthermore, inhibition of proteasomal function with MG132 (Lee and Goldberg 1998) or lactacystin (Csizmadia et al. 2010) counteracted the degradation caused by RD-43. Combined inhibition of the two pathways enhanced this effect, suggesting that both lysosomal and proteasomal pathways were involved in RD-43-mediated PTPRD degradation (Fig. 4A). Similar results were observed using immunofluorescence to detect PTPRD degradation (Supplemental Fig. S6A). Cotreatment with mAb RD-43MS and BafA1 resulted in a cytoplasmic distribution of PTPRD that overlapped with the lysosomal marker (Supplemental Fig. S6B), in which RD-43 colocalized with PTPRD (Supplemental Fig. S6C). In contrast, PTPRD rescued by treatment with proteasomal inhibitors retained a membrane-associated pattern (Supplemental Fig. S6A).
Figure 4.
RD-43 promoted PTPRD degradation via lysosomal and proteasomal pathways but independently of secretase cleavage. (A) Immunoblot analysis of CAL51 cells preincubated with vehicle (Veh), bafilomycin A1 (BafA1; 20 nM, 4 h), MG132 (5 μM, 4 h), hydroxychloroquine (HCQ; 100 μM, 4 h), and lactacystin (Lacta; 20 μM, 4 h) and treated with RD-43 (100 nM, 2 h). (Lane 1) PTPRD knockout cells (RD KO) were loaded as negative control. (B) Schematic illustration of the secretase-dependent degradation of PTPRD. Proteolytic S2 cleavage is catalyzed by α-secretase and generates an extracellular soluble ectodomain (Shedding); S3 cleavage is catalyzed by γ-secretase and generates a cytosolic soluble fragment (Intracellular cleavage) that serves as the target for proteasomal degradation. (C) Immunoblot analysis of lysates or conditioned media from 293T cells transfected with empty vector (EV) or PTPRD and treated with vehicle (Veh), TPA (1 μM, 1 h), RD-43 (100 nM, 2 h), and GI-254023X (GI; 5 μM, 4 h). (S1) The C-terminal product of S1 cleavage, (S2) the C-terminal product of S2 cleavage, (S3) the C-terminal product of S3 cleavage. (D) Immunoblot analysis of lysates of 293T cells stably expressing empty vector (EV) or PTPRD and treated with vehicle (Veh), TPA (1 μM, 1 h), RD-43 (100 nM, 2 h), DAPT (500 nM, 4 h), and lactacystin (Lacta; 20 μM, 4 h). (E) Representative confocal immunofluorescence microscopy images of 293T cells stably expressing empty vector (EV) or PTPRD treated with TPA (1 μM, 1 h), lactacystin (Lacta; 20 μM, 4 h), and RD-43MS (100 nM, 2 h) followed by staining with RD-28 (red), anti-His antibody (green), and DAPI (blue). Scale bars, 20 μm. (F) Schematic illustration of the proposed PTPRD dimer complex induced by RD-43. This complex was degraded through lysosomal and proteasomal pathways.
Secretase cleavage is a critical step during the physiological processing of LAR-like RPTPs (Cornejo et al. 2021). Two secretase cleavage products, S2 and S3 (Fig. 1A), have been identified in PTPRD, which control RPTP destruction (Fig. 4B; Phillips-Mason et al. 2011). To investigate the involvement of shedding (S2) in RD-43-mediated degradation, we collected the conditioned media from cells treated with either RD-43 or TPA (12-O-tetradecanoylphorbol-13-acetate), a PKC agonist that facilitates α-secretase-mediated receptor shedding (Lichtenthaler et al. 2018). Compared with TPA, RD-43 did not cause the accumulation of PTPRD ectodomain in the conditioned media. Furthermore, RD-43-mediated PTPRD degradation was not sensitive to GI-254023X (Ludwig et al. 2005), an α-secretase inhibitor (Fig. 4C). These data suggest that PTPRD degradation promoted by RD-43 was independent of receptor shedding.
To determine whether RD-43-induced PTPRD degradation was dependent on the cleavage of the intracellular phosphatase segment (S3), we applied a γ-secretase inhibitor to cells treated with RD-43. In cells with facilitated S2 cleavage (Fig. 4D, lanes 6–9), treatment with DAPT, a potent γ-secretase inhibitor (Dong et al. 2021), blocked the S2 product from being further processed or degraded (Fig. 4D, lane 7). Proteasomal inhibition caused the accumulation of the product of S3 cleavage (Fig. 4D, lane 8), suggesting that the S3 product was the direct target for degradation in the secretase-dependent pathway. In contrast, in RD-43-treated cells (Fig. 4D, lanes 10–13), DAPT neither generated the S2 fragment nor rescued PTPRD degradation (Fig. 4D, lane 11), suggesting that RD-43-mediated PTPRD degradation was not dependent on γ-secretase activity. More importantly, in cells treated with RD-43 and lactacystin, PTPRD molecules were rescued in the form of S1 (Fig. 4D, lanes 12,13, anti-His blot), which did not serve as the target of degradation in TPA-treated cells. Consistent with the accumulation of the C-terminal fragment, the extracellular fragment was also partially rescued by proteasomal inhibition (Fig. 4D, lanes 12,13, RD-28 blot), recapitulating the observations in CAL51 cells (Fig. 4A; Supplemental Fig. S6A). Taken together, these observations suggest that RD-43-bound PTPRD underwent a distinct degradation mechanism, in which the mature PTPRD complex (after S1 cleavage) served as the direct target of degradation machineries (Fig. 4F) in which secretase activities were dispensable.
To validate further the difference between secretase-dependent and RD-43-mediated degradation pathways, we performed immunofluorescence and examined the subcellular localization of extracellular and intracellular fragments of PTPRD generated by these pathways (Fig. 4E). In cells treated with RD-43MS and proteasome inhibitor, the His-tagged fragment remained membrane-associated, rather than adopting a cytosolic distribution due to secretase cleavage (Fig. 4E, anti-His signal, green). In contrast, the ectodomain remained colocalized with the His-tagged fragment in RD-43MS-mediated destruction, unlike in TPA-treated conditions, in which the ectodomain was shed and could not be rescued (Fig. 4E, PTPRD signal, red).
Altogether, these observations underscore that RD-43-bound PTPRD followed an unconventional degradation pathway that relied on proteasomal and lysosomal activities but was independent of secretase cleavage (Fig. 4F). Our model suggests that due to the absence of secretase cleavage, antibody-bound ectodomains were capable of engaging the intracellular catalytic fragments.
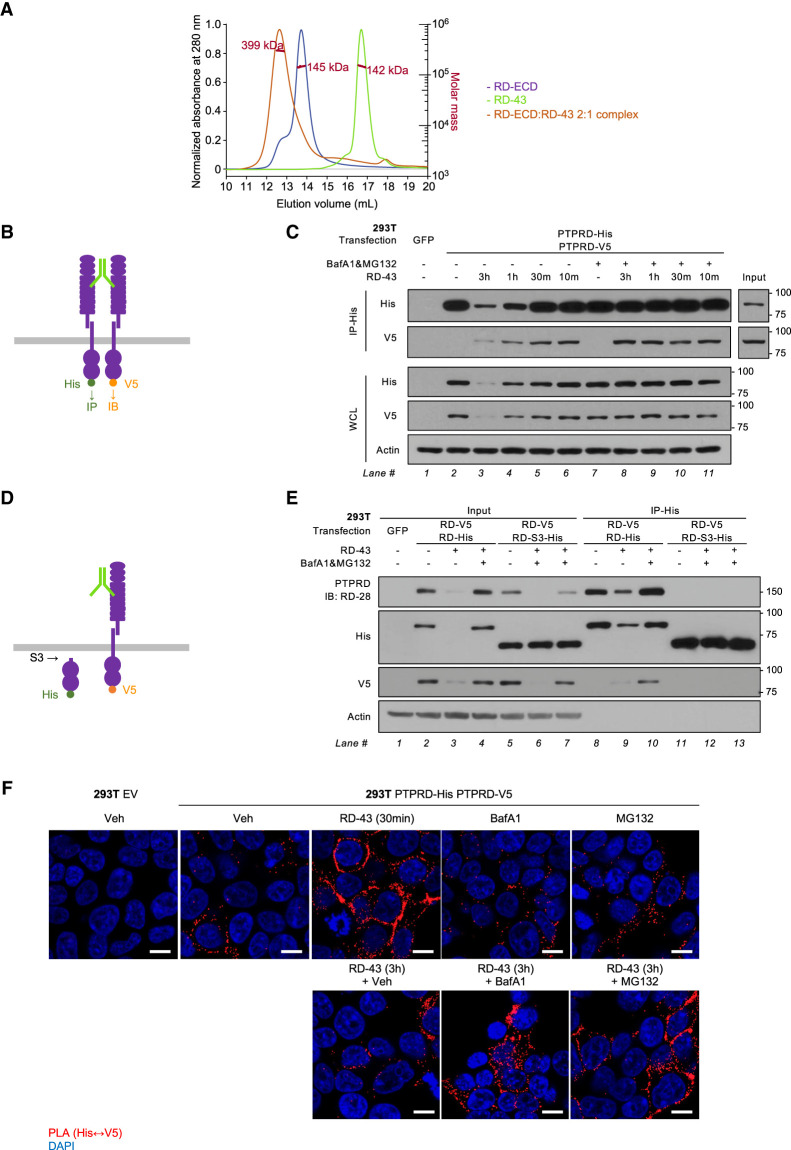
RD-43 induced dimerization of PTPRD
To assess the binding ratio between RD-43 and PTPRD ectodomain in solution, we performed size-exclusion chromatography with in-line multiangle light scattering (SEC-MALS). Similar approaches have been used to determine the monomer/dimer status of PTPRS (Coles et al. 2011). Following SEC-MALS, we concluded that RD-43 bound PTPRD ectodomain in a 1:2 molar ratio, indicated by a single peak corresponding to the antibody–antigen complex (Fig. 5A). Consistent with this, a 1:1 ratio of RD-43 to ectodomain did not saturate mAb binding, whereas a 1:4 ratio displayed unbound ectodomain (Supplemental Fig. S7).
Figure 5.
RD-43 induced dimerization of PTPRD. (A) Size-exclusion chromatography with in-line multiangle light scattering (SEC-MALS) of RD-43 alone (green), PTPRD ectodomain (RD-ECD; purple) and a 2:1 molar ratio mixture of RD-ECD and RD-43 (orange). (B) Schematic illustration of the His- and V5-tagged full-length PTPRD dimerized by RD-43. (C) Immunoblot analysis of 293T whole-cell lysate (WCL) or immunoprecipitates with anti-His antibody-conjugated beads (IP-His). 293T cells were transfected with GFP (2 μg) or 1 μg of His-tagged and 1 μg of V5-tagged PTPRD plasmid. Transfected cells were preincubated with bafilomycin (BafA1; 20 nM, 3 h) and MG132 (5 μM, 3 h) and then treated with RD-43 (100 nM) for the indicated times. (D) Schematic illustration of the V5-tagged full-length PTPRD bound by RD-43 and the His-tagged truncated PTPRD that lacks the ectodomain and transmembrane motif. (E) Immunoblot analysis of 293T whole-cell lysate (WCL) or immunoprecipitates with anti-His antibody-conjugated beads (IP-His). 293T cells were transfected with GFP (2 μg), 1 μg of V5-tagged, and 1 μg of His-tagged PTPRD (RD-V5 RD-His), or 1 μg of V5-tagged PTPRD and 1 μg of His-tagged truncated PTPRD (RD-V5 RD-S3-His) plasmids. Transfected cells were preincubated with bafilomycin (BafA1; 20 nM, 4 h) and MG132 (5 μM, 4 h) and treated with RD-43 (100 nM, 3 h). (F) Representative confocal immunofluorescence microscopy images of 293T cells expressing empty vector (EV) or PTPRD treated with vehicle (Veh), 100 nM RD-43 for the indicated durations, bafilomycin (BafA1; 20 nM, 4 h), and MG132 (5 μM, 4 h). Proximity ligation assay (PLA) signal between His and V5 tags is displayed in the red channel. Scale bars, 20 μm.
To demonstrate the formation of an antibody–PTPRD dimer complex in cells, we performed a coimmunoprecipitation assay in 293T cells expressing His- and V5-tagged PTPRD. First, we validated that His- or V5-tagged PTPRD was expressed at similar levels when individually transfected to 293T cells (Supplemental Fig. S8). Then, we investigated whether precipitation of His-tagged PTPRD could enrich V5-tagged PTPRD in the presence of RD-43 antibody (Fig. 5B). The data illustrate that short-term treatment with RD-43 was sufficient to induce interaction between His- and V5-tagged PTPRD (Fig. 5C, lane 6), suggesting that the acute SRC inhibition observed in CAL51 cells (Fig. 3B) occurred concomitantly with the rapid formation of PTPRD dimers. Upon extended treatment, in which PTPRD degradation took place, blocking lysosomal and proteasomal pathways sustained the interaction between His- and V5-tagged PTPRD (Fig. 5C, lanes 8–11; Supplemental Fig. S8).
To determine whether dimer formation was dependent on the extracellular epitope of RD-43, we tested for coprecipitation of V5-tagged full-length PTPRD and the His-tagged intracellular fragment of PTPRD (RD-S3) (Fig. 5D). Despite the presence of antibody or protein degradation inhibitors, we did not detect an interaction between the PTPRD-S3 fragment and full-length PTPRD (Fig. 5E). We conclude that through bivalent binding of the ectodomains of two PTPRD molecules, RD-43 induced the formation of PTPRD dimers that also incorporate their intracellular catalytic domains.
Finally, to demonstrate that RD-43 induced PTPRD dimerization in living cells, we performed a proximity ligation assay (PLA) to visualize the distribution of PTPRD homodimers in the prescence of protein degradation inhibitors. In 293T cells expressing His- and V5-tagged PTPRD, RD-43 triggered the interaction between the His- and V5-tagged proteins in cellular membrane regions (Fig. 5F, 30 min), consistent with the immunoprecipitation data (Fig. 5C). Coincident with PTPRD degradation during extended treatment, the PLA signal was depleted at 3 h of treatment (Fig. 5F, 3 h) but was rescued at distinct subcellular regions through lysosomal or proteasomal inhibition (Fig. 5F, BafA1 and MG132). Taken together, we showed RD-43 induced PTPRD dimerization in living cells and validated our model that RD-43-bound PTPRD dimer involved both extracellular and intracellular fragments, concomitantly serving as the target of lysosomal and proteasomal pathways (Fig. 4F).
Antibody-mediated PTPRD dimerization suppressed PTPRD-dependent invasion of breast cancer cells.
Having shown that RD-43 induced PTPRD dimerization, inhibited SRC signaling, and promoted PTPRD degradation (Fig. 3), we compared various additional PTPRD mAbs with the isotype control. Under short-term treatment, we observed that RD-43 potently down-regulated SRC phosphorylation, with two other mAbs displaying a similar, but lesser, effect compared with the isotype control (Fig. 6A,B). During long-term treatment, PTPRD mAbs facilitated protein degradation. In particular, treatment with high concentration of RD-43 led to a decrease in PTPRD protein similar to that in the genetic knockout (Fig. 6C,D). Consistent with these results, we showed that mAb RD-31 and RD-75 induced formation of PTPRD dimers to a lesser extent than RD-43 in cells (Supplemental Fig. S9), further supporting an association between the induction of PTPRD dimers and the inhibition and degradation of PTPRD.
Figure 6.
Bivalent antibody RD-43 inhibited SRC activity and PTPRD-dependent cell invasion. (A) Immunoblot analysis of CAL51 cells treated with 100 nM IgG, RD-31, and RD-75 and 25 nM or 100 nM RD-43 for 1 h. Cells treated with 50 nM dasatinib were loaded as positive controls. (B) Quantification of densitometry of SRC pTyr416/SRC calculated from three replicate immunoblots as conducted in A. (***) P-value < 0.001, (**) P-value < 0.01. (C) Immunoblot analysis of CAL51 cells treated with 100 nM IgG, RD-31, and RD-75 and 25 nM or 100 nM RD-43 overnight (O/N). PTPRD knockout cells (RD KO) were included as positive controls. (D) Quantification of densitometry of PTPRD/actin calculated from three replicate immunoblots as conducted in C. (***) P-value < 0.001. (E) Transwell assay analysis of CAL51 cells treated with 100 nM IgG, RD-31, RD-75, RD-43 (25 nM or 100 nM), or 50 nM dasatinib. Cell invasion was determined by crystal violet staining intensity normalized to IgG-treated cells. Representative images of the invasion membrane of each group are shown (n = 4). (F) Transwell assay analysis of CAL51-CAS9-sgSCB cells (sgSCB) treated with 100 nM IgG or 50 nM dasatinib, and CAL51-CAS9-sgPTPRD cells (sgRD2) treated with 100 nM IgG, RD-31, RD-43, or RD-75. Cell invasion was determined by crystal violet staining intensity normalized to IgG-treated CAL51-CAS9-sgSCB cells. Representative images of the invasion membrane of each group are shown (n = 3). For E and F, data are presented as mean ± SEM. Statistical significance was calculated using a two-tailed Student's t-test. (**) P-value < 0.01, (***) P-value < 0.001, (n/s) P-value > 0.05.
RPTP-dependent SRC activity has been reported to be essential for the invasive growth of cancer cells (Spring et al. 2015; Lin et al. 2020). To test whether RD-43-mediated PTPRD inhibition and degradation were sufficient to suppress cancer cell invasion, we assessed the invasion of CAL51 cells in transwell assays. CAL51 cells were trypsinized from tissue culture plates and resuspended in growth media for a temporary suspension culture to recover PTPRD protein levels (Supplemental Fig. S10). Cells were then resuspended in serum-free media, counted, and seeded with PTPRD mAbs in the upper chamber of transwell assay plates (Supplemental Fig. S10C). We demonstrated that, in a dose-dependent manner, RD-43 significantly reduced cell invasion, phenocopying treatment with SRC inhibitor (Fig. 6E). Moreover, we showed that PTPRD knockout cells displayed limited cell invasion and that treatment with PTPRD mAbs did not exert further inhibition (Fig. 6F). Together, these data illustrate that treatment with PTPRD-dimerizing mAbs was sufficient to suppress PTPRD-dependent invasion in CAL51 cells.
Discussion
Since the first FDA approval of a protein kinase inhibitor, imatinib, for treatment of chronic myelogenous leukemia, the number of drugs targeting various kinases across multiple cancer subtypes has increased dramatically (Rivera-Torres and San José 2019). In fact, kinase inhibitors represent a major component of modern cancer therapy and a multibillion-dollar industry. Nevertheless, drug resistance, both acquired and intrinsic, remains a major challenge to the effectiveness of kinase inhibitors in the clinic. One of the potential approaches to overcome drug resistance is combination therapy. Dual inhibition of SRC and receptor kinases, for example, has shown synergistic anticancer effects in preclinical models and is now in pharmaceutical development (Belli et al. 2020). Additionally, targeted protein degradation, including PROTAC technology (Zhao et al. 2022), has emerged as a cutting-edge approach for the destruction of specific disease-associated proteins, either alone or in combination. Our work presents an example of using a function-modulating antibody not only to inhibit enzymatic catalysis but also to facilitate the degradation of a potential therapeutic target.
In this study, we focused on a receptor tyrosine phosphatase, PTPRD, that was previously shown to promote metastasis in breast cancer models (Chaudhary et al. 2015; Yuwanita et al. 2015). We generated an array of monoclonal antibodies and extensively validated their recognition of PTPRD through various experimental methods. We demonstrated that antibody-mediated dimerization of PTPRD leads to the acute inhibition of PTPRD activity and facilitates receptor degradation following extended treatment. We uncovered a degradational pathway of mAb-bound PTPRD dimers that relies on lysosomal and proteasomal activities but does not involve extracellular shedding or intracellular cleavage by secretases. More importantly, we demonstrated that mAb RD-43 suppresses SRC activity and PTPRD-dependent invasion in a breast cancer cell model, highlighting that these antibodies may provide a new strategy for therapeutic intervention in PTPRD-dependent tumors.
The classic view that PTPs perform solely negative regulatory roles in signal transduction has been replaced by a vision of integrated signaling networks that are controlled by both protein tyrosine kinases and phosphatases. Members of the PTP family can function both negatively (to counteract PTK activity) and positively (to promote signaling events). A canonical mechanism underlying RPTP-mediated positive effects on signaling is by activation of SRC family kinases. Multiple RPTPs are capable of exerting tumor-promoting effects in certain cell contexts, such as PTPRA (Truffi et al. 2014) and PTPRJ, and have become therapeutic targets in cancer therapy; however, the dual function of such RPTPs remains to be characterized across varied cellular contexts. For example, although siRNAs targeting PTPRJ were shown to suppress cancer cell migration, invasion, and metastasis by inhibiting SRC signal (Spring et al. 2015), an inhibitory antibody targeting PTPRJ has conversely been shown to promote cell growth and angiogenesis (Takahashi et al. 2006), with such tumor suppressor effects mediated through dephosphorylation of receptor tyrosine kinases (Bloch et al. 2019). Therefore, application of such antibodies in a therapeutic context would require careful selection and validation of an appropriate patient population.
RPTPs may be regulated by dimerization-mediated inhibition, which provides a therapeutic avenue to manipulate their function through targeting their cognate ectodomains. Advantages of antibody-based pharmaceuticals include high affinity and specificity. Furthermore, the intrinsic bivalency of antibody molecules may promote dimerization; such antibodies have proven capable of reversing cellular phenotypes mediated by PTPRS (Wu et al. 2017) and PTPRJ (Takahashi et al. 2006). However, there has not been a detailed biochemical investigation regarding the fate of the dimerized RPTP molecules, particularly in the case of function-modulating antibodies. In this study, we performed extensive validation of antibody binding to the ectodomain, including dimer formation in solution in vitro (Fig. 5A), in cell lysates (Fig. 5B), and in living cells (Fig. 5F). Importantly, our data suggest that PTPRD degradation was associated with dimerization, either induced by bivalent antibody binding or chemically induced by the DmrB system. Under physiological conditions, ligand binding has also been suggested to manipulate LAR-like RPTP activity through regulation of the monomer/dimer status (Coles et al. 2011). In fact, we demonstrated that HSPG, a bivalent/multivalent ligand of PTPRS (Aricescu et al. 2002), induced PTPRS degradation in a dose-dependent manner (Supplemental Fig. S11B,C), suggesting that dimerization-associated receptor degradation may serve as a regulatory mechanism across the LAR-RPTP subfamily.
Here, we reported a secretase-independent degradational pathway for bivalent antibody-induced destruction of PTPRD. Mechanistically speaking, this degradation involved both lysosomal and proteasomal activity but was independent of secretase clevage. Ubiquitination of the intracellular segment of PTPRD, which associates noncovalently with the extracellular segment, was observed following antibody-mediated dimerization (Supplemental Fig. S11A), as well as ligand-induced dimerization of PTPRS (Supplemental Fig. S11D). Nevertheless, secretase cleavage remains a critical process during the physiological degradation of membrane proteins, including RPTPs (Phillips-Mason et al. 2011; Cornejo et al. 2021). Antibodies can facilitate the shedding of membrane proteins; this is reported to compromise the efficacy of antibody-based therapy due to the loss of epitopes in cancer cells and antibody neutralization due to accumulation of the shed ectodomain in the tumor microenvironment (Pak et al. 2014). Furthermore, ligands (Ni et al. 2001) or antibodies (Ancot et al. 2012) that bind to the extracellular segment can also promote intracellular cleavage to generate a phosphatase fragment that is catalytically active (Haapasalo et al. 2007) but no longer membrane-associated, which can bypass dimerization induced extracellularly. In this study, we observed a secretase-independent degradation pathway for RD-43-bound PTPRD. We demonstrated that the antibody–PTPRD dimer complex that formed rapidly after antibody binding remained associated during the process of degradation. This is consistent with a model in which bivalent antibodies lock PTPRD molecules in a catalytically impaired state, in which neither the extracellular fragment (which the antibodies bind to) nor the intracellular fragment (which performs the catalytic function) can escape before degradation.
Previous research demonstrated that loss of MTSS1 resulted in the up-regulation of PTPRD in breast epithelial cell models (Chaudhary et al. 2015), associated with enhanced SRC activity, and facilitated metastatic growth. Further studies should investigate whether combined anti-SRC and anti-PTPRD therapy will improve outcomes in this specific breast cancer subgroup. In addition, it is important to note that apart from the aberrant up-regulation in metastatic breast cancer, PTPRD is physiologically expressed in the central nervous system (CNS), regulates neurogenesis (Tomita et al. 2020) and synapse formation (Nakamura et al. 2017), and has been implicated in neurological diseases (Drgonova et al. 2015). Genomic studies revealed that PTPRD levels are associated with vulnerability to addiction, such as smoking and drug abuse (Drgonova et al. 2015). Indeed, PTPRD knockout mice, including heterozygous knockouts, showed significant reduction in cocaine-conditioned place preference, indicating the potential of targeting PTPRD in addiction treatment (Drgonova et al. 2015; Uhl et al. 2018). Another recent study showed that PTPRD knockout animals develop resistance to hormone-induced obesity (Mishra et al. 2022). These discoveries expand the repertoire of potential applications for targeting PTPRD in disease.
In addition to SRC, another substrate of PTPRD, STAT3, has been characterized in glioblastoma (Veeriah et al. 2009; Ortiz et al. 2014) and melanoma (Solomon et al. 2008), as well as in the overexpression system of 293T cells (Supplemental Fig. S4; Veeriah et al. 2009), and has been associated with its tumor suppressor functions. In some cases of breast cancer, miRNA has also been reported to regulate PTPRD expression and STAT3 phosphorylation (Zhang et al. 2020). It is worth noting, however, that PTPRD knockout animals do not develop spontaneous tumors (Drgonova et al. 2015; Nakamura et al. 2017). Therefore, it is important to determine the functional specificity of PTPRD in distinct contexts, especially from the perspective of targeting oncogenic PTPRD signaling specifically in cancer therapy. To that end, our work provides reliable antibody tools to assess the expression of PTPRD through multiple experimental methods. Furthermore, we have validated an inhibitory antibody that enabled rapid and specific suppression of endogenous PTPRD in genetically unmodified systems. Having now generated and mechanistically characterized monoclonal, inhibitory antibody tools for PTPRD, future studies will explore the bioactivity of these antibodies in vivo and exploit targeting RPTP signaling in a variety of disease contexts.
Materials and methods
Tissue culture, transfection, and stable cell lines
HEK293T cells (293T) were purchased from ATCC (CRL-3216). CAL51 cells are from the Cold Spring Harbor Laboratory (CSHL) Tissue Culture Facility (https://www.cshl.edu/research/core-facilities/tissue-culture). All cell lines used in this study were genetically verified and tested mycoplasma-negative. Cells were propagated in Gibco DMEM high-glucose HEPES (Thermo Fisher) with 0.5% penicillin/streptomycin (Thermo Fisher) and 10% FBS (Thermo Fisher) and, for CAL51 cells, 10% GlutaMAX supplement (Thermo Fisher).
Transient transfections in 293T cells were conducted by Trans-IT-293T transfection reagent (Mirus). For the purpose of antigen production, 15 μg of plasmid was transfected into 6.0 × 106 cells in a 10-cm plate. At 8 h posttransfection, 10 mL of culture media was changed and collected at 36 and 60 h posttransfection. For virus production, 10 μg of backbone plasmids, 5 μg of pVSVG, and 3.75 μg of pPAX (Addgene) were transfected into 2.0 × 106 cells in a six-well plate. At 8 h posttransfection, the media was changed, and viral media was collected at 36 and 60 h posttransfection. For cell signaling studies, 1.0 × 106 cells were seeded in 12-well plates. Transfection doses are specified in the figure legends.
CAL51 cells were seeded in six-well plates at 0.2 × 106 cells/well 18 h prior to infection with viral particle-containing media in the presence of 8 μg/mL polybrene. At 24 h postinfection, the media was aspirated and infected cells were incubated with complete growth media for 12 h, followed by antibiotic selection with 2 μg/mL puromycin for 48 h or 1 mg/mL G418 selection for 7 d to establish CRISPR KO cell lines or CRISPR rescue cell lines expressing gRNA-resistant PTPRD constructs. For cell signaling studies, 1.6 × 106 cells were seeded into six-well plates 24 h prior to lysis.
Production of PTPRD ectodomain
PTPRD ectodomain (ECD) residues 21–1265 from the PTPRD long isoform (isoform 1) were chosen as the template for protein production in the 293T expression system. The signal peptide derived from the human IL2 gene (MYRMQLLSCIALSLALVTNS) was fused to the N terminus of PTPRD ECD, and one His6 tag was fused to the C terminus of the construct. Purified His-tagged PTPRD ECD was purchased from ACROBiosystems.
Generation of anti-PTPRD monoclonal antibodies
Three 6-wk-old Sprague Dawley rats (Taconics) were immunized with 1.5 mg of purified PTPRD ECD (100 μg per animal per boost for five weekly boosts). Immune response was monitored by ELISA to measure the serum anti-PTPRD IgG titer from blood samples. After a 60-d immunization course, the rat with the strongest anti-PTPRD immune response was terminated, and 107 splenocytes were collected for making hybridomas by fusing with the rat myeloma cell line YB2/0, following standard methods (Greenfield 2014). All procedures were approved by the CSHL Institutional Animal Care and Use Committee (IACUC). Antibodies were purified from hybridoma culture medium by using Protein G Sepharose (GE Healthcare) according to the manufacturer's instructions.
Enzyme-linked immunoassay (ELISA)
Purified proteins were immobilized at 50 ng/well in a 384 ELISA plate (Thermo Fisher Scientific 464718) as per the manufacturer's instructions. In brief, protein-coated wells were blocked with PBS with 1% BSA overnight at 40°C prior to incubation with primary antibody. Primary and secondary antibodies were diluted in PBS-T with 1% BSA. Specified concentrations of primary antibodies described in the figures were used for binding by incubating the plates for 1 h at room temperature. Species-specific HRP-conjugated secondary antibodies (Jackson ImmunoResearch 112-035-071, 115-035-071, and 115-035-072) were used at 1:5000 dilution, and the plates were incubated for 30 min at room temperature. Three washes with PBS-T were performed between each antibody incubation step and before enzymatic signal development. Signals were developed by using TMB substrate (Thermo Fisher Scientific 34022) as per the manufacturer's instructions.
Murinization of RD-43
To engineer the chimeric antibody RD-43MS, the rat mAb V domains from RD-43 heavy and light chains were cloned onto the mouse IgG2a constant region. The resulting chimeric IgG2a antibody therefore comprised rat V domains in-frame with the constant regions of mouse IgG2a. The recombinant chimeric antibodies were then produced from 293T cells transfected with the chimeric IgG2a expression constructs, followed by purification from the culture supernatants by standard methods using Protein G Sepharose (GE Healthcare).
Generation of RD-43MS-Fab
To generate RD-43MS-Fab, purified RD-43MS was incubated with immobilized Ficin-agarose resin (Thermo Fisher Scientific 44881) in the presence of 10 mM cysteine following the manufacturer's user manual. After the Ficin digestion, the soluble phase was collected and the undigested IgG and Fc fragments were next removed from the desired Fab fragment using Protein A Sepharose. After Protein A incubation, the Fab fragment was collected from the supernatant phase followed by dialysis in PBS.
Immunoblot
Cells were washed with 1 mL of PBS and incubated with 150 μL/300 μL (12-well/six-well plate) of lysis buffer (25 mM Tris at pH 7.4, 150 mM NaCl, 1% NP-40, 5% glycerol) containing proteinase and phosphatase inhibitor cocktails (Thermo Fisher) for 30 min on ice. Lysates were then transferred to 1.5-mL microcentrifuge tubes and centrifuged at 15,000 rpm for 15 min at 4°C. Supernatants were transferred to ice-cold 1.5-mL tubes and the protein concentration was determined by Bradford assay (Bio-Rad). Lysates were then diluted to a final concentration of 2 μg/μL and mixed with 5× SDS loading buffer (5% β-mercaptoethanol, 0.02% Bromophenol blue, 30% glycerol, 10% SDS, 250 mM Tris at pH 6.8). Cell lysate (20 μL) was subjected to 10% SDS-PAGE followed by transfer to nitrocellulose membranes (GE Health). Membranes were blocked (Pierce Protein-Free T20 blocking buffer, Thermo Fisher) for 1 h at room temperature, incubated with primary antibody overnight at 4°C, incubated with secondary antibody for 1 h at 4°C (Pierce Western Blot signal enhancer, Thermo Fisher), and developed (Pierce ECL Western blotting substrate, Thermo Fisher).
Immunoprecipitation
Whole-cell lysates were produced as described above and diluted to the final concentration of 1 mg/mL. For immunoprecipitation of PTPRD, 500 μL of cell lysate was incubated with PTPRD monoclonal antibodies (4 μg/mL) for 1 h at 4°C and incubated with 25 μL of Protein G magnetic beads (Thermo Fisher), followed by the same washing steps as described below. For pull-down of His-tagged proteins, 500 μL of cell lysate was incubated with 25 μL of Ni-NTA magnetic beads (Cell Signaling Technology) overnight at 4°C and washed three times with 250 μL of PBS for 15 min at room temperature.
Flow cytometry
293T cells were seeded and transfected in low-attachment six-well plates as described above. Transfected cells were washed from the plates with PBS, centrifuged at 300g for 5 min, and resuspended in ice-cold presorting buffer (BD Biosciences). After counting, 1.5 × 105 cells were resuspended in 2 mL of presorting buffer containing PTPRD monoclonal antibody (4 μg/mL) for 30 min at 4°C, followed by fluorescent-conjugated secondary antibody (Jackson ImmunoResearch) for 15 min at 4°C. Stained cells were analyzed by LSR Dual Fortessa cell analyzer (BD Bioscience).
Immunofluorescence microscopy
For 293T cells, 0.3 × 106 cells were seeded in collagen-coated, eight-well chamber slides (Ibidi) and incubated for 24 h. For CAL51 cells, 0.6 × 106 cells were seeded in IBI-treated eight-well chamber slides (Ibidi). Cells were fixed with 4% PFA (Cell Signaling Technology) for 15 min at room temperature and washed three times with PBS. The last round of PBS washing contained 0.1% Triton X-100 (Sigma) for permeabilization. Cells were then blocked with 5% normal goat serum (Cell Signaling Technology) diluted in PBS and incubated with primary antibody overnight at 4°C followed by fluorescent secondary antibody for 1 h at 4°C. Immunofluorescence images were taken with a LSM710 confocal microscope (Zeiss).
Proximity ligation assay (PLA)
Transfected 293T cells were seeded in collagen-coated eight-well chamber slides (Ibidi) and incubated for 24 h. Cells then underwent the same process of fixation, permeabilization, blocking, and primary antibody incubation as for immunofluorescence staining. After the removal of primary antibody, cells were incubated with PLA probe-conjugated secondary antibodies (Sigma Duolink PLA fluorescence probe) for 1 h at 37°C and incubated with PLA ligation buffer for 1 h at 37°C followed by PLA amplification for 2 h at 37°C (Sigma Duolink PLA). Immunofluorescence images were gathered using a LSM710 confocal microscope (Zeiss).
Transwell assay
CAL51 cells were trypsinized, centrifuged, and resuspended in complete growth media and incubated for 4 h. Cells were centrifuged, resuspended in FBS-free media, and counted. Cells (2 × 105) were seeded in the upper chambers of Matrigel invasion transwell plates (Corning) with antibodies or dasatinib and incubated for 18 h. Residual cells were removed and those cells that had undergone invasion were fixed with ice-cold methanol for 3 min, followed by 5% crystal violet staining (Sigma) for 2 min. Images of the stained cells were taken with an ECHO Revolve microscope.
SEC-MALS
Lyophilized PTPRD-ECD protein was resuspended in water at a concentration of 1 mg/mL. The protein was then repurified using a Superdex75 Increase 10/300 column equilibrated in 10 mM Tris (pH 7.4) and 50 mM NaCl. Peak fractions were pooled, concentrated to 1–2 mg/mL, and then taken for subsequent biophysical analyses.
Analytical gel filtration was used to assess the behavior of RD-ECD, RD-43, and complexes thereof to assess initial indications of protein folding and oligomeric state. Between 4 and 10 µg of protein was applied to a Superose6 Increase 3.2/300 column at a flow rate of 0.05 mL/min in 10 mM Tris (pH 7.4) and 50 mM NaCl. Protein elution was monitored at 280 nm.
Multiangle light scattering was used to determine the oligomeric state of the RD-ECD alone and in complex with monoclonal antibodies. Protein (∼40 µg) was taken for in-line size-exclusion chromatography on a Superose6 Increase 10/300 GL column (monitored at 280 nm) followed by light scattering analysis. Chromatography was performed in a buffer of 10 mM Tris (pH 7.4) and 50 mM NaCl. MALS was measured with a Wyatt Dawn Heleos-II and processed using the included software (Astra version 5.3.4). Bovine serum albumin (BSA) was used as calibration standard.
Statistics
All results are expressed as the mean ± SEM. ANOVA and a two-tailed Student's t-test were used to determine statistical significance. A P-value of ≤0.05 was considered significant. Statistical analysis and generation of graphs were performed using GraphPad Prism version 9 (GraphPad Software).
Supplementary Material
Acknowledgments
N.K.T. is the Caryl Boies Professor of Cancer Research at Cold Spring Harbor Laboratory (CSHL). Research in the Tonks laboratory was supported by National Institutes of Health grants CA53840 and DK124907 and CSHL Cancer Center Support Grant CA45508, in addition to support from the Robertson Research Fund of CSHL, the Don Monti Memorial Research Foundation, the Irving A. Hansen Memorial Foundation, and the Simons Foundation. Research in the Yeh laboratory was supported by CSHL Cancer Center Support Grant CA45508, the Robertson Research Fund of CSHL, and the Cold Spring Harbor Laboratory and Northwell Health Affiliation. L.J.-T. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: Z.Q., D.S., J.J.I., J.T.-H.Y., and N.K.T. designed experiments. Z.Q. performed experiments. D.S. analyzed the data of flow cytometry and generated the protocol for PLA. C.B. and J.T.-H.Y. conducted purification and murinization of the antibodies. J.J.I .and L.J.-T. conducted and analyzed SEC-MALS. Z.Q. and N.K.T. wrote the manuscript, and all authors contributed to editing. J.T-.H.Y. and N.K.T. conceptualized and supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350713.123.
Competing interest statement
N.K.T. serves on the science advisory board of DepYmed, Inc., and Anavo Therapeutics.
References
- Aicher B, Lerch M, Müller T, Schilling J, Ullrich A. 1997. Cellular redistribution of protein tyrosine phosphatases LAR and PTPσ by inducible proteolytic processing. J Cell Biol 138: 681–696. 10.1083/jcb.138.3.681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancot F, Leroy C, Muharram G, Lefebvre J, Vicogne J, Lemiere A, Kherrouche Z, Foveau B, Pourtier A, Melnyk O, et al. 2012. Shedding-generated Met receptor fragments can be routed to either the proteasomal or the lysosomal degradation pathway. Traffic 13: 1261–1272. 10.1111/j.1600-0854.2012.01384.x [DOI] [PubMed] [Google Scholar]
- Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. 2006. Furin-, ADAM 10-, and γ-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of β-catenin's transcriptional activity. Mol Cell Biol 26: 3917–3934. 10.1128/MCB.26.10.3917-3934.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, McKinnell IW, Halfter W, Stoker AW. 2002. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase σ. Mol Cell Biol 22: 1881–1892. 10.1128/MCB.22.6.1881-1892.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli S, Esposito D, Servetto A, Pesapane A, Formisano L, Bianco R. 2020. c-Src and EGFR inhibition in molecular cancer therapy: what else can we improve? Cancers 12: 1489. 10.3390/cancers12061489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, den Hertog J, Hunter T, Noel JP. 1996. Structural basis for inhibition of receptor protein-tyrosine phosphatase-α by dimerization. Nature 382: 555–559. 10.1038/382555a0 [DOI] [PubMed] [Google Scholar]
- Bloch E, Sikorski EL, Pontoriero D, Day EK, Berger BW, Lazzara MJ, Thévenin D. 2019. Disrupting the transmembrane domain-mediated oligomerization of protein tyrosine phosphatase receptor J inhibits EGFR-driven cancer cell phenotypes. J Biol Chem 294: 18796–18806. 10.1074/jbc.RA119.010229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, Sloan AE, Miller RH, Basilion JP, Brady-Kalnay SM. 2010. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase μ. Neoplasia 12: 305–316. 10.1593/neo.91940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary F, Lucito R, Tonks NK. 2015. Missing-in-metastasis regulates cell motility and invasion via PTPδ-mediated changes in SRC activity. Biochem J 465: 89–101. 10.1042/BJ20140573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-NP, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH-T, Chen Z, Cooke VG, et al. 2016. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535: 148–152. 10.1038/nature18621 [DOI] [PubMed] [Google Scholar]
- Chin C-N, Sachs JN, Engelman DM. 2005. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett 579: 3855–3858. 10.1016/j.febslet.2005.05.071 [DOI] [PubMed] [Google Scholar]
- Coles CH, Shen Y, Tenney AP, Siebold C, Sutton GC, Lu W, Gallagher JT, Jones EY, Flanagan JG, Aricescu AR. 2011. Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science 332: 484–488. 10.1126/science.1200840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo F, Cortés BI, Findlay GM, Cancino GI. 2021. LAR receptor tyrosine phosphatase family in healthy and diseased brain. Front Cell Dev Biol 9: 659951. 10.3389/fcell.2021.659951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csizmadia V, Csizmadia E, Silverman L, Simpson C, Raczynski A, O'Brien L, Gallacher M, Cardoza K, Kadambi VJ, Fedyk ER, et al. 2010. Effect of proteasome inhibitors with different chemical structures on the ubiquitin-proteasome system in vitro. Vet Pathol 47: 358–367. 10.1177/0300985809358423 [DOI] [PubMed] [Google Scholar]
- Dong Z, Huo J, Liang A, Chen J, Chen G, Liu D. 2021. γ-Secretase inhibitor (DAPT), a potential therapeutic target drug, caused neurotoxicity in planarian regeneration by inhibiting Notch signaling pathway. Sci Total Environ 781: 146735. 10.1016/j.scitotenv.2021.146735 [DOI] [PubMed] [Google Scholar]
- Drgonova J, Walther D, Wang KJ, Hartstein GL, Lochte B, Troncoso J, Uetani N, Iwakura Y, Uhl GR. 2015. Mouse model for PTPRD associations with WED/RLS and addiction: reduced expression alters locomotion, sleep behaviors and cocaine-conditioned place preference. Mol Med 21: 717–725. 10.2119/molmed.2015.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvel B, Yasri A. 2014. Antibodies directed against receptor tyrosine kinases: current and future strategies to fight cancer. MAbs 6: 838–851. 10.4161/mabs.29089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa A, Sugawara H, Tanga N, Ishii K, Kuboyama K, Uchiyama S, Suzuki R, Noda M. 2019. A head-to-toe dimerization has physiological relevance for ligand-induced inactivation of protein tyrosine receptor type Z. J Biol Chem 294: 14953–14965. 10.1074/jbc.RA119.007878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada M, Fujikawa A, Chow JPH, Ikematsu S, Sakuma S, Noda M. 2006. Protein tyrosine phosphatase receptor type Z is inactivated by ligand-induced oligomerization. FEBS Lett 580: 4051–4056. 10.1016/j.febslet.2006.06.041 [DOI] [PubMed] [Google Scholar]
- Greenfield EA. 2014. Preface. In Antibodies: a laboratory manual, 2nd ed. (ed. Greenfield EA), pp. xxi. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Haapasalo A, Kim DY, Carey BW, Turunen MK, Pettingell WH, Kovacs DM. 2007. Presenilin/γ-secretase-mediated cleavage regulates association of leukocyte–common antigen-related (LAR) receptor tyrosine phosphatase with β-catenin. J Biol Chem 282: 9063–9072. 10.1074/jbc.M611324200 [DOI] [PubMed] [Google Scholar]
- Haque A, Andersen JN, Salmeen A, Barford D, Tonks NK. 2011. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell 147: 185–198. 10.1016/j.cell.2011.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2015. Discovering the first tyrosine kinase. Proc Natl Acad Sci 112: 7877–7882. 10.1073/pnas.1508223112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Barford D, Flint AJ, Tonks NK. 1995. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science 268: 1754–1758. 10.1126/science.7540771 [DOI] [PubMed] [Google Scholar]
- Kennedy LC, Gadi V. 2018. Dasatinib in breast cancer: Src-ing for response in all the wrong kinases. Ann Transl Med 6: S60. 10.21037/atm.2018.10.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584: 1393–1398. 10.1016/j.febslet.2009.12.047 [DOI] [PubMed] [Google Scholar]
- Krebs EG. 1993. Protein phosphorylation and cellular regulation I (Nobel lecture). Angew Chem Int Ed Engl 32: 1122–1129. 10.1002/anie.199311221 [DOI] [Google Scholar]
- Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss C-M, Page R, Blackledge M, et al. 2014. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol 10: 558–566. 10.1038/nchembio.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BT, Cregg JM, DePaul MA, Tran AP, Xu K, Dyck SM, Madalena KM, Brown BP, Weng Y-L, Li S, et al. 2015. Modulation of the proteoglycan receptor PTPσ promotes recovery after spinal cord injury. Nature 518: 404–408. 10.1038/nature13974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8: 397–403. 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF, Lemberg MK, Fluhrer R. 2018. Proteolytic ectodomain shedding of membrane proteins in mammals—hardware, concepts, and recent developments. EMBO J 37: e99456. 10.15252/embj.201899456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Xin S, Huang X, Zhang F. 2020. PTPRA facilitates cancer growth and migration via the TNF-α-mediated PTPRA–NF-κB pathway in MCF-7 breast cancer cells. Oncol Lett 20: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R-M, Hwang Y-C, Liu I-J, Lee C-C, Tsai H-Z, Li H-J, Wu H-C. 2020. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 27: 1. 10.1186/s12929-019-0592-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD. 2005. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen 8: 161–171. 10.2174/1386207053258488 [DOI] [PubMed] [Google Scholar]
- Majeti R, Bilwes AM, Noel JP, Hunter T, Weiss A. 1998. Dimerization-induced inhibition of receptor protein tyrosine phosphatase function through an inhibitory wedge. Science 279: 88–91. 10.1126/science.279.5347.88 [DOI] [PubMed] [Google Scholar]
- Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema K-J, Coppes RP, Engedal N, Mari M, Reggiori F. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14: 1435–1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilahti JAM, Elenius K. 2019. γ-Secretase-dependent signaling of receptor tyrosine kinases. Oncogene 38: 151–163. 10.1038/s41388-018-0465-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra I, Xie WR, Bournat JC, He Y, Wang C, Silva ES, Liu H, Ku Z, Chen Y, Erokwu BO, et al. 2022. Protein tyrosine phosphatase receptor δ serves as the orexigenic asprosin receptor. Cell Metab 34: 549–563.e8. 10.1016/j.cmet.2022.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. 2018. Phosphatases start shedding their stigma of undruggability. Nat Rev Drug Discov 17: 847–849. 10.1038/nrd.2018.201 [DOI] [PubMed] [Google Scholar]
- Nakamura F, Okada T, Shishikura M, Uetani N, Taniguchi M, Yagi T, Iwakura Y, Ohshima T, Goshima Y, Strittmatter SM. 2017. Protein tyrosine phosphatase δ mediates the Sema3A-induced cortical basal dendritic arborization through the activation of Fyn tyrosine kinase. J Neurosci 37: 7125–7139. 10.1523/JNEUROSCI.2519-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. 2001. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294: 2179–2181. 10.1126/science.1065412 [DOI] [PubMed] [Google Scholar]
- Ortiz B, Fabius AWM, Wu WH, Pedraza A, Brennan CW, Schultz N, Pitter KL, Bromberg JF, Huse JT, Holland EC, et al. 2014. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci 111: 8149–8154. 10.1073/pnas.1401952111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak Y, Pastan I, Kreitman RJ, Lee B. 2014. Effect of antigen shedding on targeted delivery of immunotoxins in solid tumors from a mathematical model. PLoS One 9: e110716. 10.1371/journal.pone.0110716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Mason PJ, Craig SEL, Brady-Kalnay SM. 2011. Should I stay or should I go? Shedding of RPTPs in cancer cells switches signals from stabilizing cell-cell adhesion to driving cell migration. Cell Adh Migr 5: 298–305. 10.4161/cam.5.4.16970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido R, Krueger NX, Serra-Pagès C, Saito H, Streuli M. 1995. Molecular characterization of the human transmembrane protein-tyrosine phosphatase δ. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase δ isoforms. J Biol Chem 270: 6722–6728. 10.1074/jbc.270.12.6722 [DOI] [PubMed] [Google Scholar]
- Rivera-Torres J, San José E. 2019. Src tyrosine kinase inhibitors: new perspectives on their immune, antiviral, and senotherapeutic potential. Front Pharmacol 10: 1011. 10.3389/fphar.2019.01011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih I-M, Wang T-L. 2007. Notch signaling, γ-secretase inhibitors, and cancer therapy. Cancer Res 67: 1879–1882. 10.1158/0008-5472.CAN-06-3958 [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim J-S, Cronin JC, Sibenaller Z, Ryken T, Rosenberg SA, Ressom H, Jean W, Bigner D, Yan H, et al. 2008. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res 68: 10300–10306. 10.1158/0008-5472.CAN-08-3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring K, Fournier P, Lapointe L, Chabot C, Roussy J, Pommey S, Stagg J, Royal I. 2015. The protein tyrosine phosphatase DEP-1/PTPRJ promotes breast cancer cell invasion and metastasis. Oncogene 34: 5536–5547. 10.1038/onc.2015.9 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Takahashi K, Mernaugh RL, Tsuboi N, Liu H, Daniel TO. 2006. A monoclonal antibody against CD148, a receptor-like tyrosine phosphatase, inhibits endothelial-cell growth and angiogenesis. Blood 108: 1234–1242. 10.1182/blood-2005-10-4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tertoolen LG, Blanchetot C, Jiang G, Overvoorde J, Gadella TW, Hunter T, den Hertog J. 2001. Dimerization of receptor protein-tyrosine phosphatase α in living cells. BMC Cell Biol 2: 8. 10.1186/1471-2121-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Cornejo F, Aranda-Pino B, Woodard CL, Rioseco CC, Neel BG, Alvarez AR, Kaplan DR, Miller FD, Cancino GI. 2020. The protein tyrosine phosphatase receptor δ regulates developmental neurogenesis. Cell Rep 30: 215–228.e5. 10.1016/j.celrep.2019.11.033 [DOI] [PubMed] [Google Scholar]
- Tonks NK. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 7: 833–846. 10.1038/nrm2039 [DOI] [PubMed] [Google Scholar]
- Tonks NK. 2013. Protein tyrosine phosphatases—from housekeeping enzymes to master regulators of signal transduction. FEBS J 280: 346–378. 10.1111/febs.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffi M, Dubreuil V, Liang X, Vacaresse N, Nigon F, Han SP, Yap AS, Gomez GA, Sap J. 2014. RPTPα controls epithelial adherens junctions, linking E-cadherin engagement to c-Src-mediated phosphorylation of cortactin. J Cell Sci 127: 2420–2432. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Martinez MJ, Paik P, Sulima A, Bi G-H, Iyer MR, Gardner E, Rice KC, Xi Z-X. 2018. Cocaine reward is reduced by decreased expression of receptor-type protein tyrosine phosphatase D (PTPRD) and by a novel PTPRD antagonist. Proc Natl Acad Sci 115: 11597–11602. 10.1073/pnas.1720446115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijk T, Blanchetot C, den Hertog J. 2005. Regulation of receptor protein-tyrosine phosphatase dimerization. Methods 35: 73–79. 10.1016/j.ymeth.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J, et al. 2009. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci 106: 9435–9440. 10.1073/pnas.0900571106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Yang S, Wakabayashi K, Svensson MND, Stanford SM, Santelli E, Bottini N. 2020. RPTPα phosphatase activity is allosterically regulated by the membrane-distal catalytic domain. J Biol Chem 295: 4923–4936. 10.1074/jbc.RA119.011808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodings JA, Sharp SJ, Machesky LM. 2003. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase δ. Biochem J 371: 463–471. 10.1042/bj20021962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-L, Hardy S, Aubry I, Landry M, Haggarty A, Saragovi HU, Tremblay ML. 2017. Identification of function-regulating antibodies targeting the receptor protein tyrosine phosphatase σ ectodomain. PLoS One 12: e0178489. 10.1371/journal.pone.0178489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Weiss A. 2002. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nat Immunol 3: 764–771. 10.1038/ni822 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23: 33–42. 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravendijk MR, Dalgarno D, et al. 2000. Investigating protein-ligand interactions with a mutant FKBP possessing a designed specificity pocket. J Med Chem 43: 1135–1142. 10.1021/jm9904396 [DOI] [PubMed] [Google Scholar]
- Yeatman TJ. 2004. A renaissance for SRC. Nat Rev Cancer 4: 470–480. 10.1038/nrc1366 [DOI] [PubMed] [Google Scholar]
- Yuwanita I, Barnes D, Monterey MD, O'Reilly S, Andrechek ER. 2015. Increased metastasis with loss of E2F2 in Myc-driven tumors. Oncotarget 6: 38210–38224. 10.18632/oncotarget.5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang B, Qin T, Wang L, Zhang Q, Lu Y, Song B, Yu X, Li L. 2020. IL-6 induces tumor suppressor protein tyrosine phosphatase receptor type D by inhibiting miR-34a to prevent IL-6 signaling overactivation. Mol Cell Biochem 473: 1–13. 10.1007/s11010-020-03803-w [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhao J, Zhong K, Tong A, Jia D. 2022. Targeted protein degradation: mechanisms, strategies and application. Signal Transduct Target Ther 7: 113. 10.1038/s41392-022-00966-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.