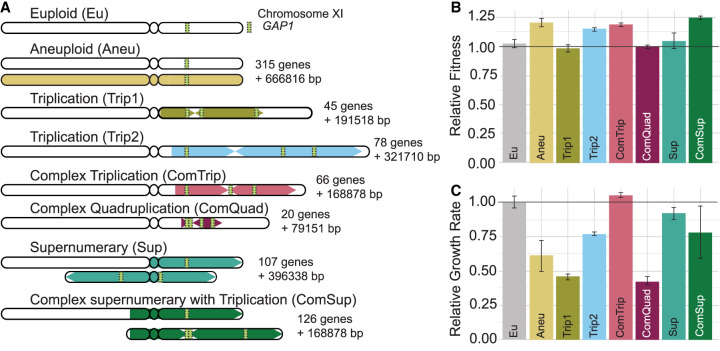
Figure 1.
Strains with GAP1 CNVs differ in structure and fitness. (A) We previously evolved a euploid Saccharomyces cerevisiae strain in glutamine-limited chemostats and isolated seven strains that have CNVs on Chr XI that include GAP1. The structure of each GAP1 CNV was resolved using long-read sequencing and is summarized here. The amplified region is shown as a colored block with arrows. Arrows pointing right represent copies that maintain their original orientation, whereas arrows pointing left represent copies that are inverted. The number of genes amplified and the number of additional base pairs (bp) are annotated. (B) The relative fitness (compared with the ancestral strain) of strains containing GAP1 CNVs was determined by pairwise competition experiments in glutamine-limited chemostats. Error bars are 95% confidence intervals for the slope of the linear regression used to compute fitness (see Methods). (C) Average and standard deviation (error bars) growth rate relative to the ancestral, euploid strain in YPGal batch culture. Horizontal black lines in B and C denote the ancestral euploid fitness. Note that panels B and C do not show the same type of measurement.