Figure 6.
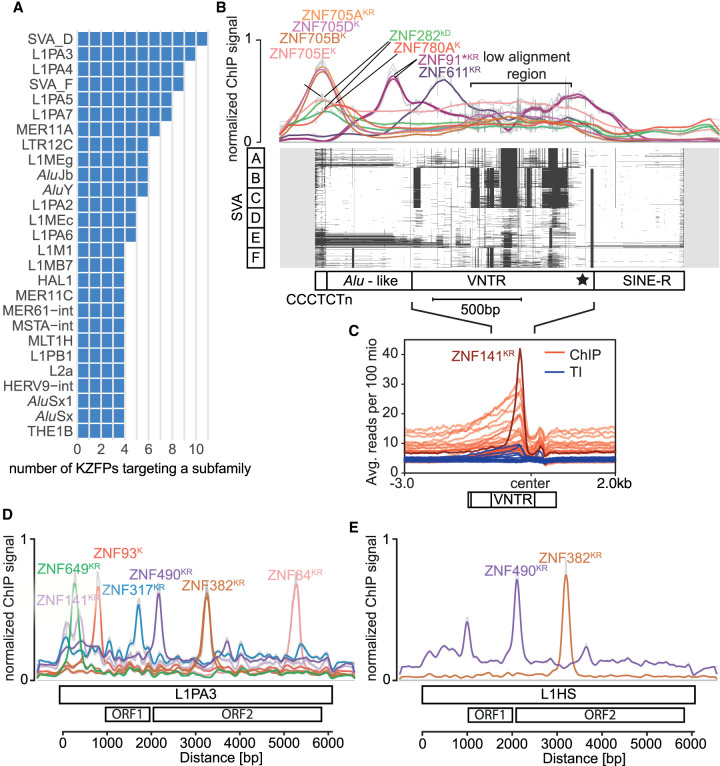
TE families are targeted by multiple KZFPs. (A) Bar graphs showing the TE subfamilies targeted by the largest number of different KZFPs. Only KZFPs targeting the subfamily as their primary targets were considered (−log10(FDR) within 10% of the highest −log10(FDR) for that KZFP). (B) KZFP signal over the multiple sequence alignment (MSA) of SVA subfamilies A to F. Top: Line graph of the normalized cumulative reads for each position from the indicated ChIP-seq and -exo experiments. External data sets are marked with stars. Bottom: MSA plot of 100 of the longest SVA sequences for each subfamily indicated on the left, 200 bp of nonaligned extensions are added around elements shown in gray, white depicts aligned regions, and black gaps in the alignment. For visibility, places in the alignment (columns) with more than 85% gaps were removed. The approximate different domains of the SVAs are indicated below, adapted from Hancks and Kazazian (2010); the star indicates the center region for C. (C) Signal over the low alignment region of the remaining SVA binders centered on the 3′ end of the VNTR (without alignment of sequences). ChIP signals for KZFPs enriched on SVAs are shown in red (ZFP57, ZFP92*, ZNF14*, ZNF141, ZNF155*, ZNF215, ZNF25, ZNF256, ZNF263, ZNF268*, ZNF28, ZNF30, ZNF41*, ZNF415*, ZNF461*, ZNF500*, ZNF556*, ZNF560*, ZNF57*, ZNF587B*, ZNF597, ZNF624*, ZNF641, ZNF689*, ZNF699*, ZNF747*, ZNF813*, ZNF852*, and ZNF878*; * = new data in this publication) with the signal for ZNF141 shown in dark red. Input signals for the presented ChIPs are shown in blue. (D,E) Binding sites of KZFPs on L1PA3 and L1HS elements. Elements were aligned the same way as in A and the normalized ChIP-seq and -exo signals are shown for each aligned position. External data sets are marked with stars. K = standard KRAB, k = variant KRAB, D = DUF domain, R = repressor; according to Helleboid et al. (2019) and Tycko et al. (2020). (D) 1000 L1PA3 elements were aligned. (E) 382 full-length L1HS elements were aligned. Multimapped reads were included for the signals in panels B–E.