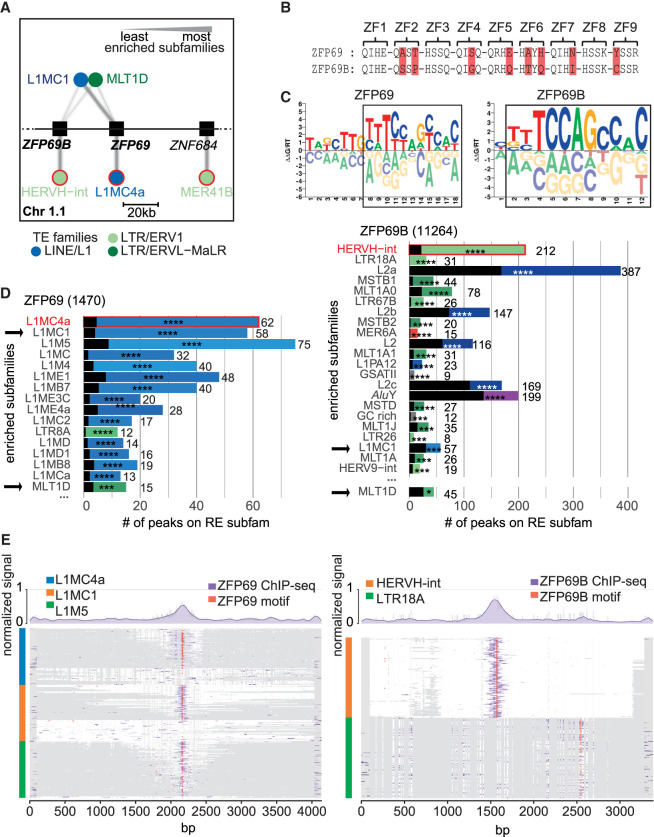
Figure 7.
Evolution of TE-KZFP interaction. (A) Network for cluster Chr 1.1 in which targets (circles) of each KZFP (squares) are shown as connected edges and the amount of binding is represented by the line thickness. The thickest line for each KZFP represents the TE subfamily with the highest −log10(FDR) and then scales linearly to the lowest value. For visibility, only the best targets (below) and shared targets (above) are shown. The TE subfamilies are colored according to their families. Primary targets for each KZFP are highlighted in red. (B) Zinc fingerprints of ZFP69 and ZFP69B. The DNA-contacting amino acids for zinc finger (ZF) are shown; differences are highlighted in red. (C) DNA binding motifs of ZFP69 and ZFP69B as identified by Weirauch et al. (2014). Regions of high similarity are framed by a black square. (D) Enrichment of peaks over different repetitive element subfamilies. Subfamilies with FDR > 0.01 are shown. The width of the colored bars represents the number of peaks per subfamily also shown as a number on the right of the bar. The black transparent bars represent the expected number of peaks following a random distribution. The FDR of the enrichment is shown with stars (FDR > 0.0001 = ****, >0.001 = ***, >0.01 = **, >0.05 = *, ≥0.05 = n.s.). Rows are ordered by FDR. The number next to the title indicates the total number of peaks for the experiment. Primary targets for each KZFP are highlighted in red and shared subfamilies between the two panels are indicated by black arrows. (E) MSA over the three most enriched targets of ZFP69 (left) and ZFP69B (right). Up to 200 elements for the indicated targets (blue, orange, and green) were aligned, selecting first elements overlapping with a peak and then the longest elements. White regions in the plots indicate aligned sequences; gray regions indicate gaps. The signal of ZFP69 and ZFP69B ChIPs was laid over their respective alignments in purple. The locations of their motifs from panel C are shown in red. The normalized signal can be seen as a line plot above the MSA plot.