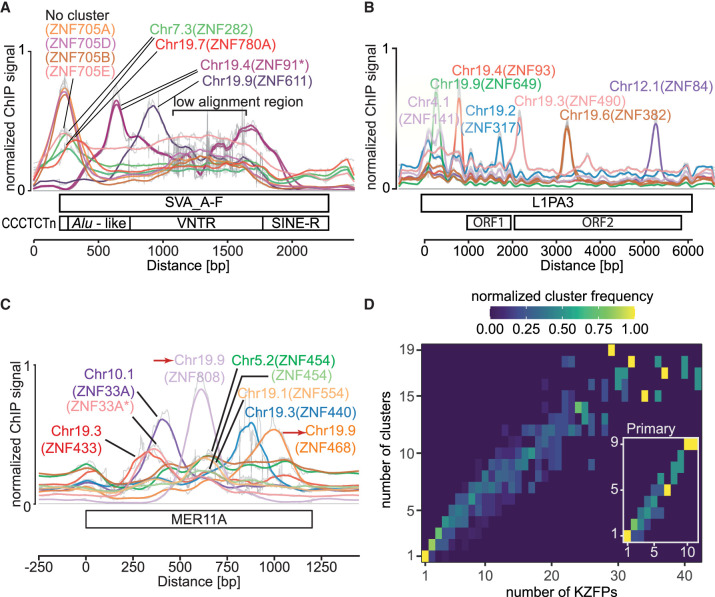
Figure 8.
Localization of multiple KZFPs targeting the same TE subfamily. (A–C) Cluster location of the indicated KZFPs, which primarily bind the respective TE subfamilies. Duplicated clusters are marked with red arrows; external data sets are marked with stars. (A) Alignment of approximately 200 of the longest SVA_A to SVA_F elements. (B,C) Alignment of 1000 L1PA3 and MER11A elements. (D) Heatmap comparing the genomic locations of KZFP genes, the products of which target the same TE subfamilies, showing that they are spread across multiple gene clusters. Each square represents the indicated number (x-axis) of different KZFPs targeting the same TE subfamily and the number (y-axis) of KZFP gene clusters in which these KZFPs are located. Colors represent the frequency with which the number of KZFPs are found in the number of different clusters and are normalized for each column. Main panel: all KZFPs targeting a subfamily (FDR > 0.05). Inset: Only KZFPs primarily targeting a subfamily (−log10(FDR) within 10% of the highest −log10(FDR)).