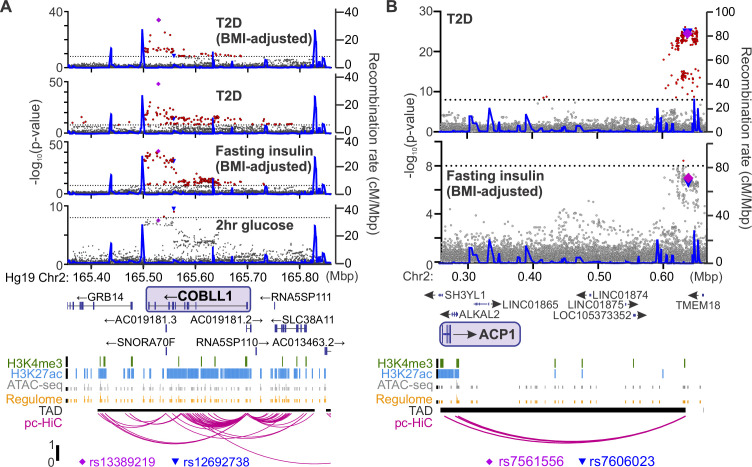
Figure 6. Identifying candidate protein targets by integrating human genome-wide association studies (GWAS).
(A) An example gene, COBLL1, orthologous to a gene coding for a protein identified as highly correlated to Ca2+ wave parameters in the founder mice. The recombination rate is indicated by the solid blue line. Significant single-nucleotide polymorphisms (SNPs; 8 < −log10(p), red) decorate the gene body for multiple glycemia-related parameters (in bold). Human islet chromatin data (Miguel-Escalada et al., 2019) for histone methylation (H3K4me3), histone acetylation (H3K27ac), ATAC-sequencing (ATAC-seq), and regulome score suggest active transcription of the gene within a topologically associated domain (TAD). Human islet promoter-capture HiC data (pc-HiC) (Miguel-Escalada et al., 2019) show contacts between the SNP-containing regions decorating the gene and its promoter. The highest SNP for 2 hr glucose (▼) and the other parameters (♦) are indicated. (B) Some orthologues did not show SNPs decorating the gene itself but did show looping to regions with SNPs for glycemic traits. The promoter of ACP1, for example, loops to a region within its topologically associated domain (black bar) with strong SNPs for type 2 diabetes risk and near-threshold SNPs for fasting insulin adjusted for body mass index (BMI). Some SNPs (▼, ♦) lie directly on the contact regions identified by HiC, whereas others lie immediately proximal to these contacts. For both panels, the significance of association (−log10 of the p-value) for the individual SNPs is on the left y axis and the recombination rate per megabasepair (Mbp) is on the right y axis. Chromosomal position in Mbp is aligned to Hg19. SNP data were provided by the Common Metabolic Diseases Knowledge Portal (https://hugeamp.org/).