Figure 2.
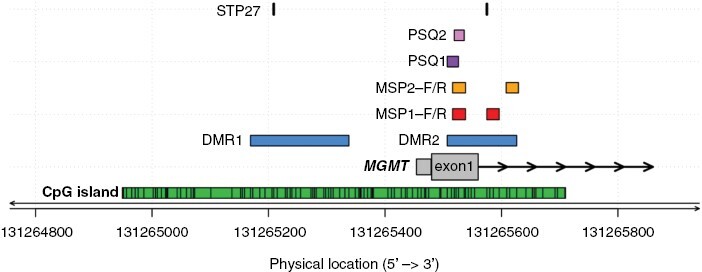
CpGs interrogated in commonly used MGMT promoter methylation assays. The physical location of sites in the CpG island (green) interrogated by commonly used MGMT methylation assays are shown (CpGs, numbered 1–98, genome build GRCh37/hg19). The differentially methylated regions 1 and 2 (DMR1 and 2) are marked in blue.74 The CpGs from the original MSP assay commonly examined by qualitative and quantitative MSP assays are marked in red (MSP1, CpGs 76–80 and 84–87);73,77,79–81 CpGs analyzed in a quantitative MSP assay (MSP2, CpGs 76–80 and 88–90) used in large clinical trials are marked in yellow.76 Two sets of CpGs used commonly for pyrosequencing, PSQ1 (CpGs 74–78) and PSQ2 (CpGs 76–79), are marked in purple and pink, respectively.77,80 The locations of the CpGs (31, 84) selected from the BeadChip array analyzed by the MGMT-STP27 procedure, are marked in black.58 Figure by Pierre Bady.
