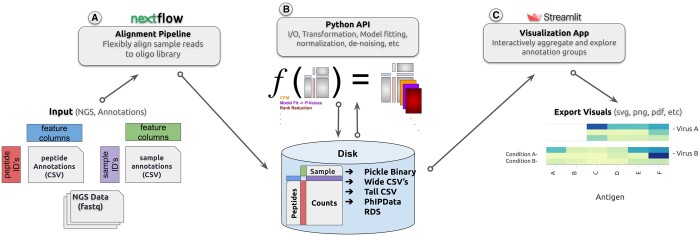
Figure 1.
(A) The processing of sequencing data from a PhIP-Seq experiment begins with the alignment pipeline, which generates either a CSV file or xarray object containing the peptide counts across samples and associated annotations. (B) Custom analysis is performed with phippery library function calls, for example via the command line interface, and (C) visualization of results can be presented in an interactive manner using our Streamlit app.