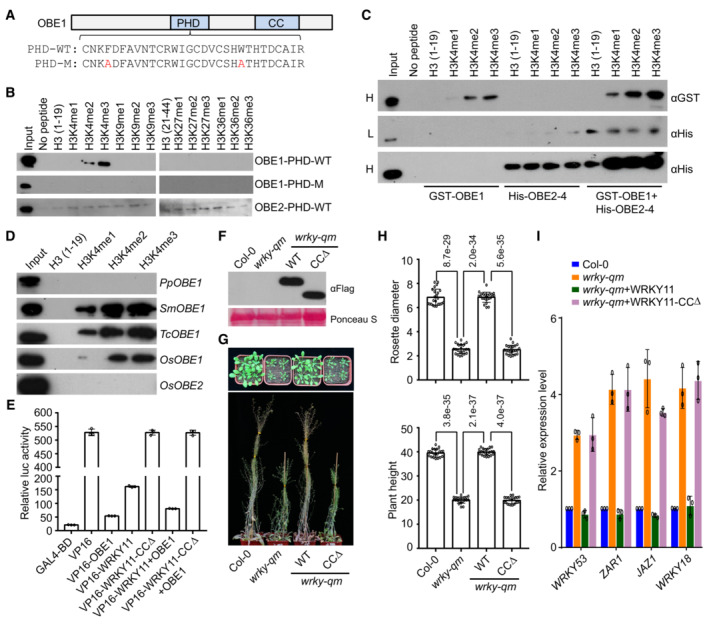
Figure 7. OBE proteins bind to histone peptides and are responsible for transcriptional repression of WRKY‐OBE complexes.
-
ADiagram of the wild‐type and mutated PHD domains in OBE1. Mutated residues in the PHD domain are shown in red.
-
BInteraction of the PHD domains of OBE1 and OBE2 with histone peptides as determined by pull‐down assays. The wild‐type and mutated PHD domains in OBE1 and the wild‐type PHD domain in OBE2 were purified and then mixed with indicated histone peptides for pull‐down assays.
-
CInteraction of GST‐OBE1, His‐OBE2, and the GST‐OBE1 and His‐OBE2 mixture with histone peptides as determined by pull‐down assays. H, high exposure; L, low exposure.
-
DInteraction of OBE1 and/or OBE2 orthologs with histone peptides as determined by pull‐down assays. PpOBE1, SmOBE1, TcOBE1, and OsOBE1 are OBE1 orthologs in Physcomitrium patens (Pp), Selaginella moellendorffii (Sm), Taxus chinensis (Tc), and Oryza sativa (Os), respectively. OsOBE2 is an OBE2 ortholog in Oryza sativa.
-
EDetermination of the transcriptional repression activity of OBE1, WRKY11, WRKY11‐CCΔ, and the mixture of WRKY11 or WRKY11‐CCΔ with OBE1 by a luciferase reporter assay. The transcriptional repression activity of OBE1, WRKY11‐WT, and WRKY11‐CCΔ was determined by fusing with the transcription factor VP16. Values are means ± SD of three independent biological replicates.
-
FThe expression levels of Flag‐tagged wild‐type WRKY11 and WRKY‐CCΔ proteins determined by immunoblotting. The ribosome protein stained by Ponceau S is indicated as a loading control.
-
G, HRestoration of the developmental defects of the wrky‐qm mutant by WRKY11 and WRKY11‐CCΔ transgenes. Morphological phenotype (G) and the statistical data of rosette diameter and plant height (H) are shown. The data were calculated from a minimum of 20 plants. Values are means ± SD. P‐values were determined by two‐tailed Student's t‐test and are indicated above columns.
-
IExpression levels of representative stress‐responsive genes in wild‐type, wrky‐qm, and WRKY11 and WRKY11‐CCΔ transgenic lines. Values are means ± SD of three biological replicates.
Source data are available online for this figure.
