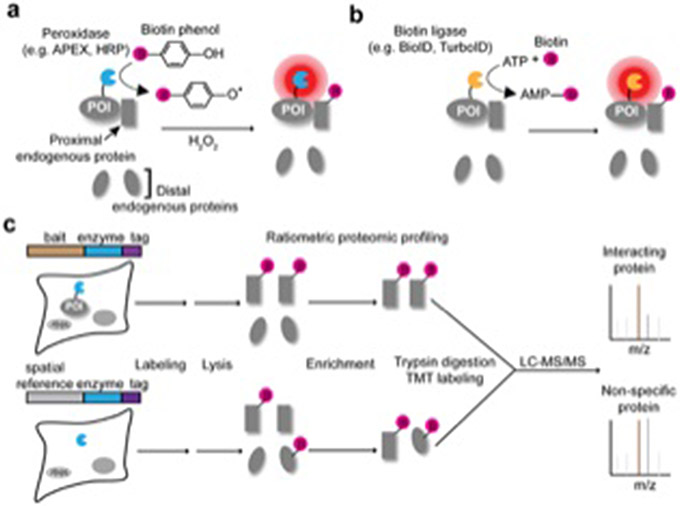
Figure 1.
Peroxidase- and biotin ligase-based proximity labeling methods for PPI mapping. (a) Peroxidase-based approaches, such as APEX or HRP, oxidize biotin phenol into reactive phenoxyl radicals using hydrogen peroxide, which preferentially labels proximal over distal endogenous proteins. (b) Biotin ligase-based approaches, such as BioID or TurboID, utilize ATP and biotin to catalyze the formation of reactive biotin-5’-AMP intermediates, which diffuse and label proximal proteins. (c) Schematic of example proteomic workflow for mapping PPI. PL enzymes fused to the bait of interest and a spatial reference control are expressed in separate samples. Biotinylated proteins from each sample are enriched and analyzed via quantitative mass spectrometry. Proteins that preferentially interact with the bait of interest can be identified by ratiometric analysis.