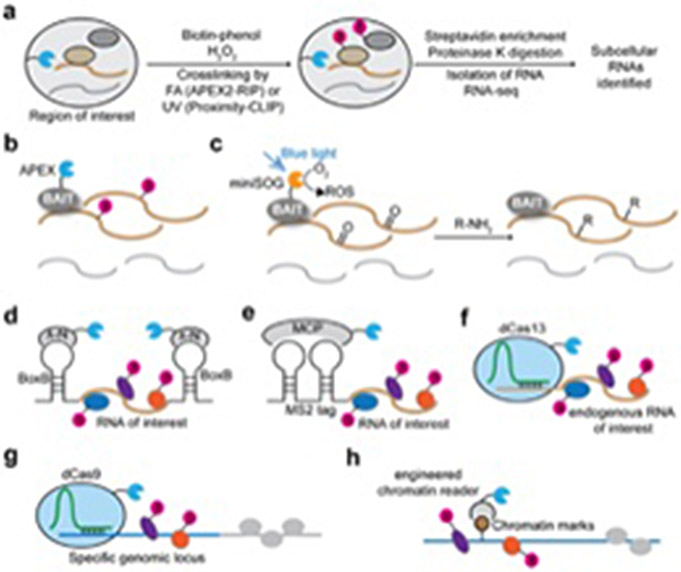
Figure 2.
PL-based methods to investigate protein-nucleic acid interactions. (a) Schematic of APEX-RIP and Proximity-CLIP. APEX targeted to a specific subcellular location catalyzes the biotinylation of proximal proteins, and the RNA-protein interactions are subsequently crosslinked by either UV or formaldehyde (FA). The subcellular RBP-occupied RNA can be captured via streptavidin-based enrichment of the biotinylated RBPs. (b) Schematic of APEX-seq. APEX directly biotinylates proximal RNA (yellow), but not distal RNA (grey), of a protein bait. (c) Schematic of Cap-seq. Upon blue light illumination, miniSOG generates ROS that react with guanine nucleobases in RNA. The photo-oxidation intermediates are intercepted by amine probes (R-NH2) to form covalent adducts. (d) Schematic of RaPID. An RNA of interest is tagged with a BoxB aptamer to recruit a fusion protein of λ-N and a promiscuous biotin ligase, which can biotinylate associated RBPs. (e) PL strategies based on MS2 tags and MCP to capture RBPs associated with an RNA of interest. (f) dCas13-based PL strategies to biotinylate RBPs associated with an endogenous RNA of interest. (g) dCas9-based PL strategies to biotinylate DNA-binding proteins at specific genomic locus. (h) Schematic of ChromID. BASU is fused to engineered chromatin readers that can specifically recognize particular chromatin marks, leading to the biotinylation of chromatin-binding proteins.