Figure 1.
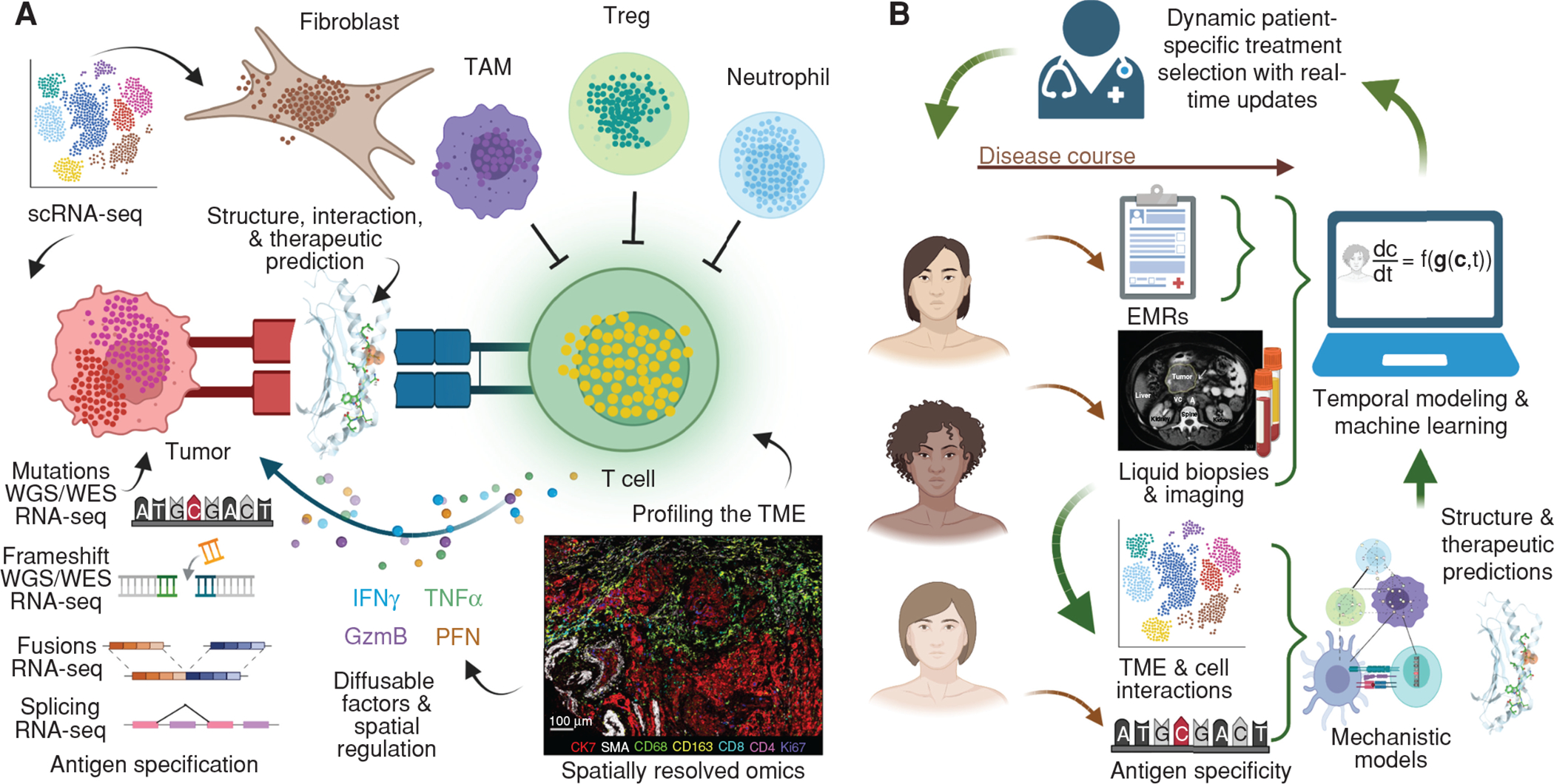
Integration of clinical, biological, technological, and mathematical expertise to design new frameworks to monitor and infer evolutionary changes in tumor and immune interactions that guide a new predictive medicine paradigm. A, Spatial and single-cell profiling of the TME over multiple time points in cancer development will allow for deeper understanding of the dynamics of immune populations and interactions with tumor cells as cancer progression occurs over time and upon intervention with therapeutics. T-cell receptor modeling with HLA–antigen complexes will also allow us to understand epitope recognition and evasion. WES/WGS/RNA-seq data will determine neoantigen evolution in tumor cells over time and upon therapeutic intervention. RNA-seq, RNA sequencing; scRNA-seq, single-cell RNA sequencing; TAM, tumor-associated macrophage; Treg, regulatory T cell; WES, whole-exome sequencing; WGS, whole-genome sequencing. B, Integration of these large datasets detailing dynamics of TME and antigenic properties of tumor cells with clinical datasets including electronic medical systems, liquid biopsies, and imaging data will allow for the development of machine learning and predictive algorithms that could guide dynamic patient-specific treatment selection in real time. EMR, electronic medical record.
