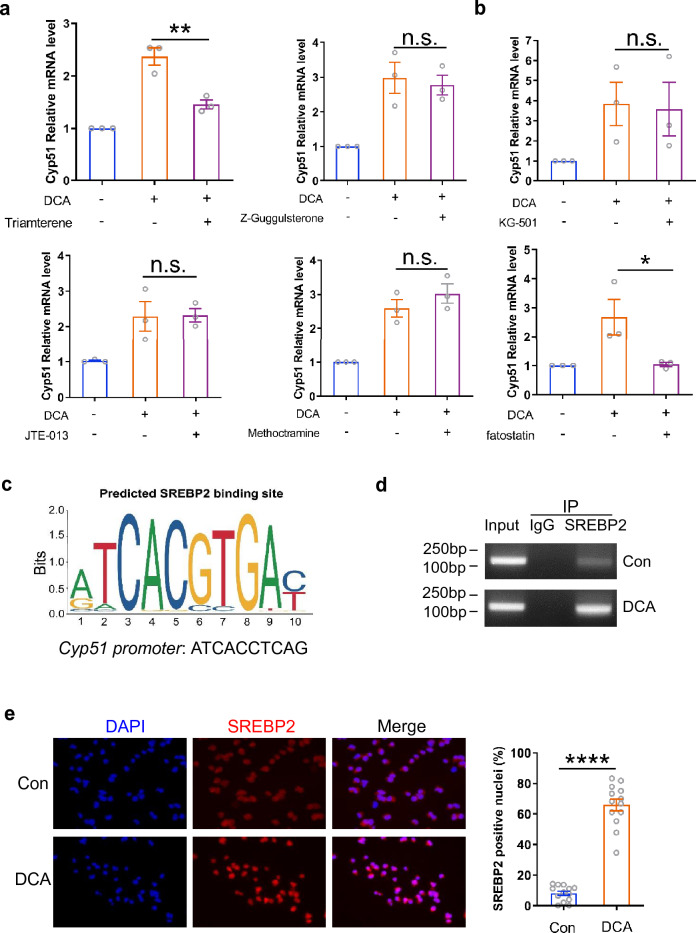
Fig. 5.
DCA increases CYP51 expression by targeting transcriptional factor SREBP2. a EL4 cells were stimulated with DCA (100 µM) in the presence or absence of Triamterene (10 μM, TGR5 inhibitor), Z-Guggulsterone (20 μM, FXR inhibitor), JTE-013 (10 μM, S1PR2 inhibitor) or methoctramine (5 µM, M2-mAchR inhibitor). The mRNA expression levels of CYP51 were determined by real-time PCR (n = 3). b EL4 cells were stimulated with DCA (100 µM) in the presence or absence of KG-501 (10 µM) or fatostatin (10 µM). The mRNA expression levels of CYP51 were determined by real-time PCR (n = 3). c Predicted SREBP2 binding site on Cyp51 promoter. Colorful letters were the motif of SREBF2 binding site cited from JASPAR. d EL4 cells were stimulated with or without DCA (100 µM) for 24 h and then ChIP assay was performed. Anti-SREBP2 antibody or isotype-matched IgG control antibody were used. PCR was applied to quantify the precipitated DNA with primers flanking the SREBP2 binding region of the CYP51 promoter. e Representative SREBP2 (red) and DAPI (blue) immunofluorescence staining of EL4 cells untreated or treated with DCA (n = 3). Data are pooled from three independent experiments. Each dot represents one microscopic high power field (HPF); four to five HPFs were scored per experiment and totally at least 300 cells were evaluated. *p < 0.05; **p < 0.01; ****p < 0.0001. n.s.: no statistically significant difference (p > 0.05). One-way ANOVA with Tukey's multiple comparisons tests (a, b) or two-tailed Student’s t-test with Welch's correction (e: right panel) was used. Data are expressed as mean ± SEM from at least three independent experiments or representative data (d, e: left panel)