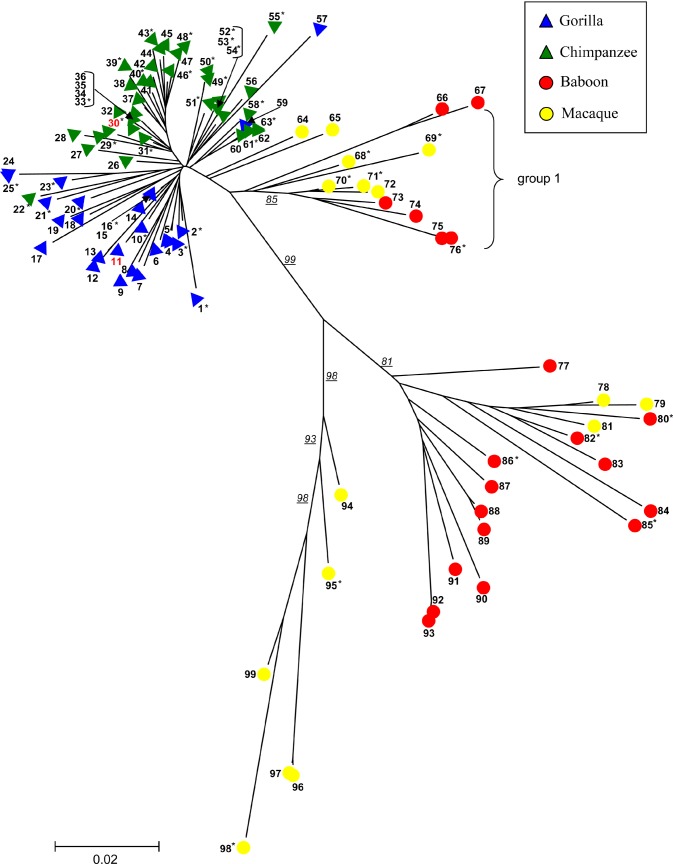
Figure 4. PTERV1 Phylogenetic Tree.
Portions of the gag and env genes (about 823 bp) were resequenced from 101 PTERV1 elements from common chimpanzee (n = 42), gorilla (n = 25), rhesus macaque (n = 14), and olive baboon (n = 20). A neighbor-joining phylogenetic tree shows a monophyletic origin for the gorilla and chimpanzee endogenous retroviruses but a polyphyletic origin among the Old World monkey species. Bootstrap support (n = 10,000 replicates) for individual branches are underlined. Although the retroviral insertions have occurred after speciation, retroviral sequences show greater divergence than expected for a non-coding nuclear DNA element (see Table S4). Table S8 provides a clone key for number designation. Phylogenetic trees showing the gag, env, and LTR segments separately are presented in Figure S6. Sequences 11 and 30 (red) are mapped to one of the 12 ambiguous overlapping loci described in the text (see Table S3). They do not cluster in this phylogenetic tree, which indicates that they are unlikely to be true orthologs.