Table 2. Cross-Species Retroviral Insertion Mapping.
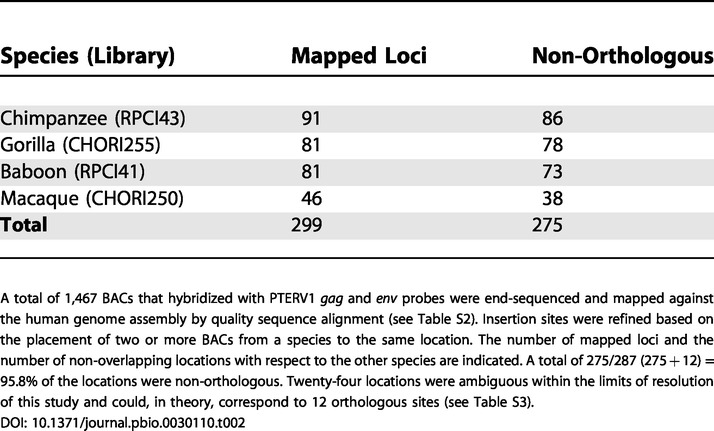
A total of 1,467 BACs that hybridized with PTERV1 gag and env probes were end-sequenced and mapped against the human genome assembly by quality sequence alignment (see Table S2). Insertion sites were refined based on the placement of two or more BACs from a species to the same location. The number of mapped loci and the number of non-overlapping locations with respect to the other species are indicated. A total of 275/287 (275 + 12) = 95.8% of the locations were non-orthologous. Twenty-four locations were ambiguous within the limits of resolution of this study and could, in theory, correspond to 12 orthologous sites (see Table S3)
