Figure 5. Tumor gland expression subtype composition and organization further suggest co-expressor cells as an intermediate state between pure classical and pure basal cells.
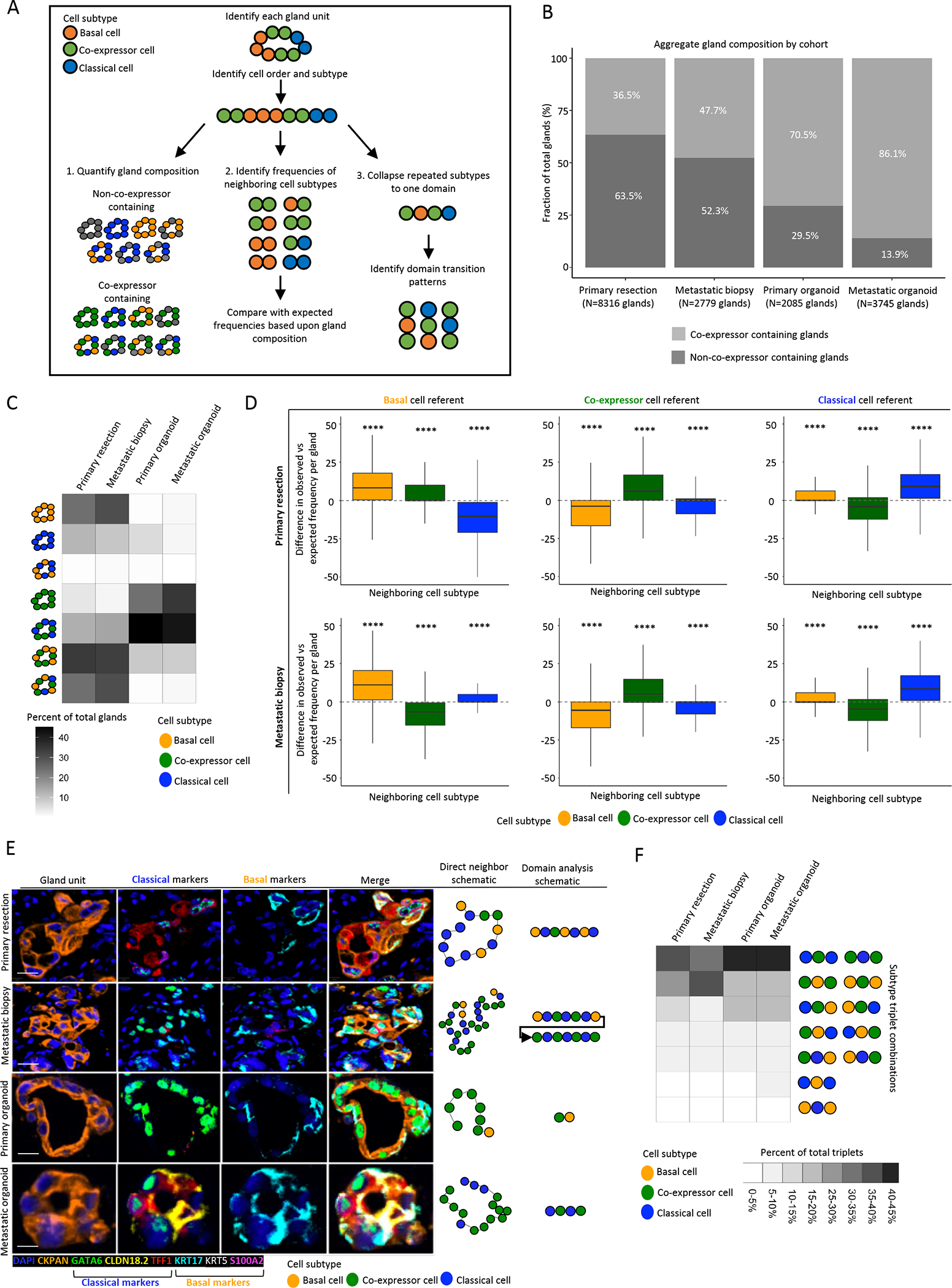
A: Schematic representing analysis workflow. Following gland-unit identification, cell order and subtype were determined. Gland composition identified, and direct neighbor and domain analysis conducted. B: Gland type distribution in primary tissue, metastatic tissue, primary organoid and metastatic organoid specimens. C: Distribution of gland composition for co-expressor containing and non-co-expressor containing glands by specimen type for gland units containing subtype informative cells, demonstrating gland units comprised of basal and classical cells rarely observed across cohorts and most gland units to be a mixture of expression subtypes. D: Direct neighbor analysis with comparison of expected and observed frequencies of subtypes by cell of reference and specimen type. Basal cells neighbor basal cells and classical cells neighbor classical cells more frequently than expected by chance. Difference between observed an expected frequencies per gland unit depicted (Mann-Whitney test between observed and expected frequencies) (Outliers removed from boxplots). E: Representative mIF images of gland-units by specimen type (top to bottom), scale bar = 10μm. Left to right: Gland-unit: DAPI and CKPAN, Classical markers: GATA6, CLDN18.2 and TFF1, Basal markers: KRT17, KRT5 and S100A2, Merge, Direct neighbor analysis schematic of cell subtypes in gland unit, Domain analysis schematic of concatenated domains of cell subtypes. E: Domain analysis of gland-units derived from primary resection, metastatic biopsy and patient-derived organoids. Domains of co-expressor cells are frequently flanked by domains of classical or basal cells, while domains of basal and classical cells are rarely directly adjacent. **** P<0.0001.
