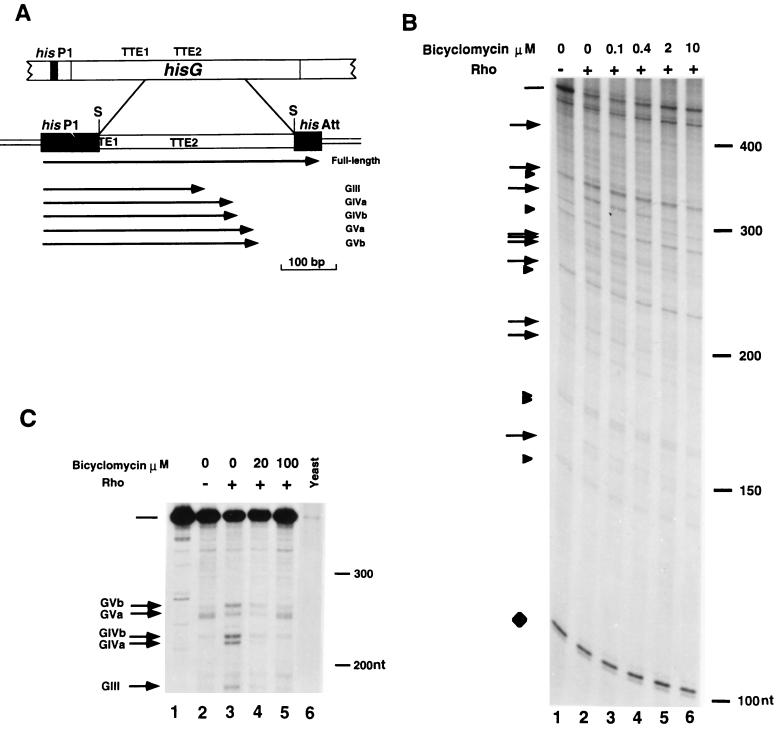
FIG. 1.
In vitro Rho-dependent transcription termination in the hisG cistron in the presence of bicyclomycin. (A) A genetic map of the promoter-proximal region of the his operon of S. typhimurium is shown at the top of panel A. The relative positions of the his P1 primary promoter, the structural hisG gene, and the intracistronic Rho-dependent TTEs (TTE1 and TTE2) are indicated. An enlarged map of the hisG region contained in plasmid pHG360 is shown at the bottom of panel A. This plasmid was obtained by cloning a 359-bp Sau3AI DNA fragment spanning the TTE2 between the his P1 primary promoter and the his attenuator (his Att) (3). The arrows identify the positions of the 3′ ends and the lengths of the major transcripts produced in vitro. S, Sau3AI. (B) Plasmid pHG360 was transcribed in vitro with E. coli RNA polymerase and [α-32P]UTP either in the absence (lane 1) or in the presence (lanes 2 to 6) of purified Rho. In lanes 3 to 6 bicyclomycin was added at a concentration of 0.1 to 10 μM. The arrows on the left indicate the positions of the putative Rho-dependent terminated transcripts. Arrowheads correspond to Rho-independent transcripts. The full-length transcript is indicated by a bar. A 104-nucleotide vector-specific Rho-independent transcript (7) is shown as a loading control (diamond). Densitometric values of the full-length transcript in the absence of Rho (lane 1) and in the presence of Rho (lane 2) or Rho plus 100 μM bicyclomycin (lane 6) were determined by normalizing the intensities to the intensity of the 104-nucleotide (nt) transcript. The positions of molecular weight markers (100, 150, 200, and 300 nucleotides) are indicated on the right. (C) S1 nuclease mapping of the transcripts in the hisG cistron terminated in vitro. Plasmid pHG360 was transcribed in vitro either in the absence (lane 2) or in the presence (lanes 3 to 5) of purified Rho. Bicyclomycin was added at concentrations of 20 μM (lane 4) and 100 μM (lane 5). The RNAs synthesized in vitro or 20 μg of yeast RNA (lane 6) were hybridized to the 3′-end labelled strand derived from the 359-bp Sau3AI fragment complementary to the RNA (lane 1). The hybrids were treated with S1 nuclease and were electrophoresed on a 5% acrylamide denaturing gel. The arrows on the left indicate the more prominent 3′ ends corresponding to Rho-dependent terminated transcripts observed previously (3). The full-length protected transcript is indicated by a bar. The positions of the molecular weight markers (200 and 300 nucleotides) are indicated on the right.