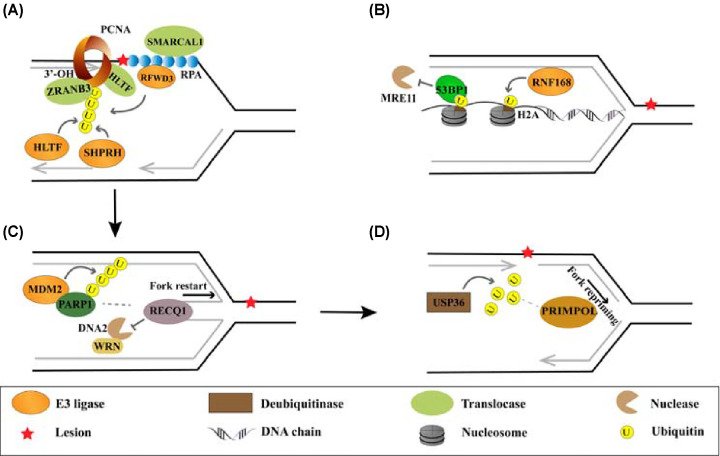
Figure 2. Regulation of fork reversal and restart by ubiquitination.
(A) Fork reversal is initiated by DNA translocases such as SMARCAL1, ZRANB3, and HLTF. SMARCAL1 binds to RPA-coated ssDNA, while ZRANB3 is recruited at stalling forks via polyubiquitinated PCNA mediated by RFWD3, HLTF, or SHPRH, and HLTF has an affinity for available 3′-OH groups located near the fork junction. These three translocases may act on different types of stalled forks or perform distinct functions at the same fork. (B) The regressed arm of a reversed fork requires protection against nucleolytic attacks. RNF168-mediated monoubiquitination of H2A on K13/15 at stalled forks is recognized by 53BP1, protecting the regressed arms from degradation by MRE11: this modification also regulates nucleosome deposition, facilitating efficient fork restart. (C) Replication fork restart or repriming requires specific factors such as RECQ1 and PRIMPOL. RECQ1 activity is increased following the ubiquitination and inactivation of PARP1 by the oncogenic E3 ligase MDM2, which accelerates DNA replication progression. (D) Deubiquitination of PRIMPOL by USP36 enhances its stability at stressed forks, contributing to replication repriming.