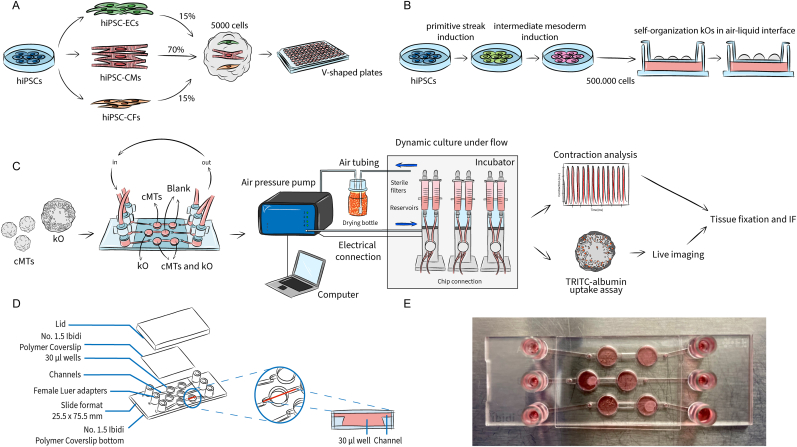
Fig. 1.
Formation of organoid models and microfluidic setup. (A) cMTs were generated using hiPSC-derived cardiomyocytes (CMs), cardiac endothelial cells (ECs) and cardiac fibroblasts (CFs). After mixing them in defined ratios, the cardiac cells self-aggregated to form cMTs and were cultured in V-shaped 96 wells plates. (B) kOs were generated from undifferentiated hiPSCs. After one week of differentiation in a monolayer, cells were detached and subjected to microcentrifugation. Pellets of 500.000 cells were then pipetted onto a membrane and cultured in an air-liquid interface. (C) Schematic representation of the workflow: after the organoid generation, the tissues were loaded into the chip and kept in dynamic culture conditions for 72 h. The integrated Ibidi setup comprises a pump system which is, on one hand, connected to the computer and controlled through the PumpControl software; inside the incubator, on the other hand, the pumps are connected to 3 FUs, where the black perfusion set is mounted and whose tubing is in turn attached to each channel of the μ-Slide III 3D Perfusion chip. Organoids were retrieved and used for functional downstream analyses, i.e. contraction analysis (for cMTs), TRITC-albumin uptake assay (for kOs) and immunostaining (both). (D) The geometry of the μ-Slide III 3D Perfusion chip is characterized by three channels where two wells are interconnected, allowing the co-culture of the two organoids while keeping them separated; the perfusion set tubing of each FU is connected to the reservoirs of the channels through its end luers; an adhesive coverslip allows for the sealing of the on-chip culture chambers; the lid aids the transportation of the loaded chip before its connection and after retrieval. (E) Image of a loaded chip.