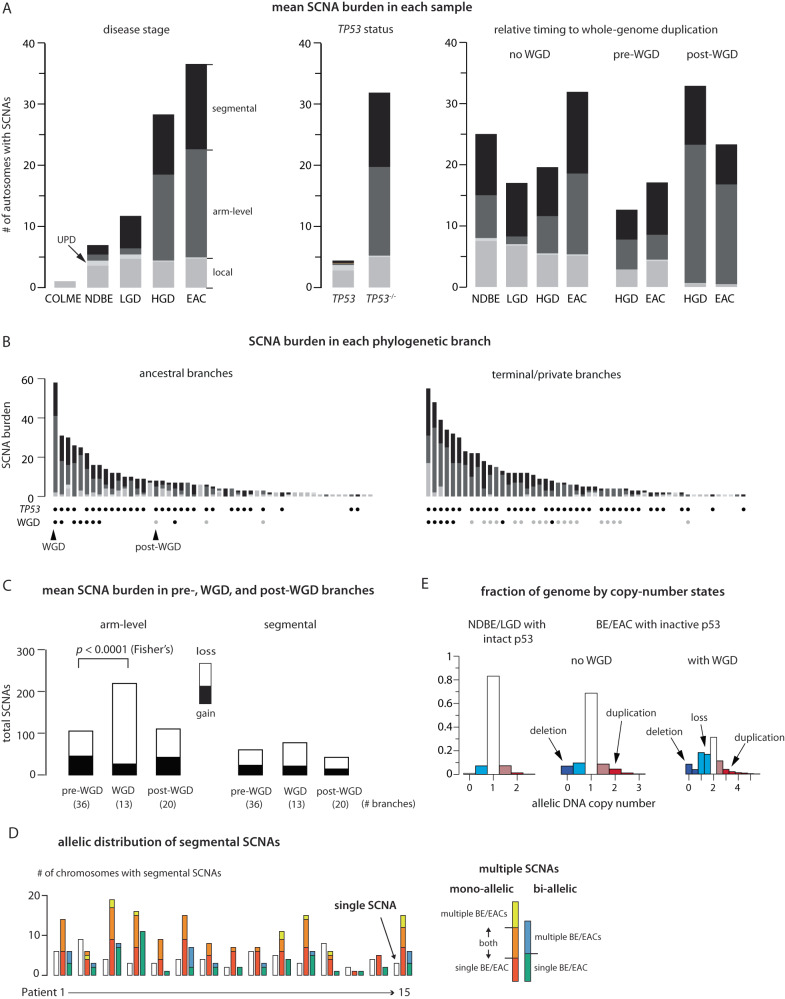
Fig. 4. Landscape of somatic copy-number alterations (SCNA) in BE and EACs.
A Mean SCNA burden in samples grouped by disease stage (left), TP53 mutation status (middle), and timing relative to whole-genome duplication (right). The SCNA burden is measured by the total number of altered autosomes (both parental homologs, maximum 44) and subdivided into local deletions or duplications (gray), uniparental disomies (light gray), arm-level SCNAs (dark gray), and segmental SCNAs (black). See Supplementary Fig. 3A–D for the SCNA burden in each sample and of subcategories of segmental SCNAs. In the middle panel, the ‘intact’ TP53 group (“TP53”) only includes NDBE/LGD samples without detectable TP53 alterations, but not HGD/EAC samples. See Supplementary Fig. 3C for the SCNA burden in HGD/EAC samples without p53 inactivation. B SCNA burden along ancestral (having more than one progeny clone) and terminal (only one progeny clone) phylogenetic branches. The bottom shows the TP53 mutation status and the relative timing to WGD of each branch. C Total counts of arm-level (left) and segmental (right) SCNAs (filled bars for gains, open bars for losses) in evolutionary branches preceding, concurrent with, or after WGD. Segmental SCNAs only include large internal/terminal SCNAs but not complex SCNAs that can generate both DNA gain and loss. The significantly higher burden of arm-level SCNAs in WGD-concurrent branches than pre-WGD branches (Mann-Whitney p = 3 × 10-6; 95% Confidence Interval: 7-16; Effect Size: 0.68) is dominated by chromosome losses (Two-sided Fisher’s test p = 10−9; 95% Confidence Interval: 0.10–0.33; Effect Size: 0.18,), consistent with chromosome losses after tetraploidization. WGD is also associated with a modest but significant increase of segmental SCNA burden (WGD-concurrent vs pre-WGD: Mann–Whitney p = 0.0071; 95% confidence interval: 1–5; effect size: 0.43) and of arm-level SCNAs (post-WGD vs pre-WGD: Mann-Whitney p = 0.0032; 95% confidence interval: 1–4; effect size: 0.40). D Allelic distribution of segmental SCNAs identified in all samples from each patient. Shown are the number of chromosomes (Chrs.1-22 and X) with single SCNAs (open bars), multiple SCNAs affecting a single parental homolog (‘mono-allelic’), or multiple SCNAs affecting both homologs (‘bi-allelic’). Mono-allelic and bi-allelic SCNAs with multiple breakpoints are further divided into subcategories based on whether SCNA breakpoints are found in a single BE/EAC genome, or in multiple related BE/EAC genomes. See Supplementary Fig. 3E. E Fraction of the germline genome at different copy-number states (from 100kb-level allelic copy number). Deletion (dark blue), subclonal deletion/loss (light blue), subclonal gain (light red), or duplication (dark red).