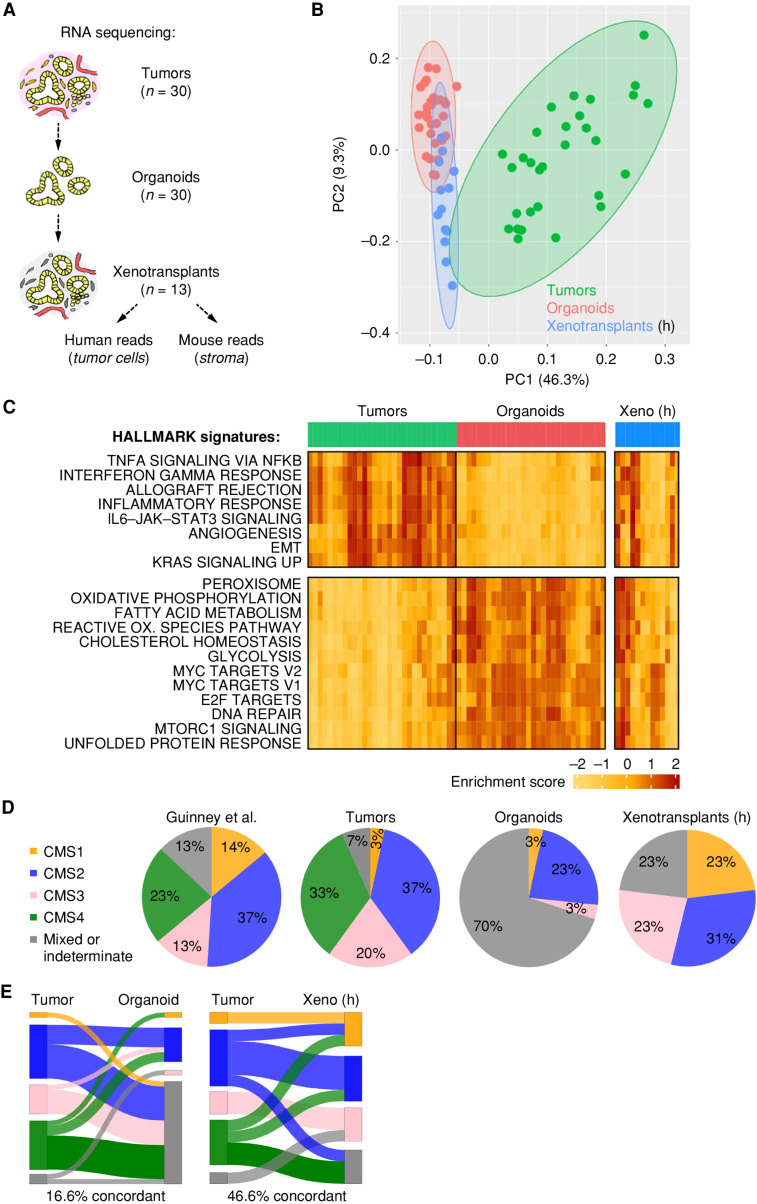
Figure 2.
Microenvironmental context is essential for the manifestation of transcriptomic subtypes. A, Schematic strategy for RNA sequencing in matched samples from primary tumors and organoids (n = 30 each), and following subcutaneous PDTO transplantation in NSG mice. An arbitrary subset of xenotransplants (n = 13) was analyzed by RNA sequencing, and human reads (h) were studied. For information on xenotransplant growth, see Supplementary Table S2. B, PCA shows transcriptomic differences among tumors and organoids and partial normalization in xenografts. C, Single-sample GSVA. Unsupervised clustering of tumors and organoids; the most differentially regulated HALLMARK signatures are shown. Note the restoration of tumor-specific signatures in xenografted organoids. EMT, epithelial-to-mesenchymal transition; OX., oxygen; Xeno, xenotransplant. D, CMS classification in clinical samples (Guinney et al.; ref. 3) and experimental models (see Supplementary Table S5). E, Sankey plots show weak overlap between tumors and organoids and increased concordance upon organoid xenotransplantation. See Supplementary Figs. S4 and S5.