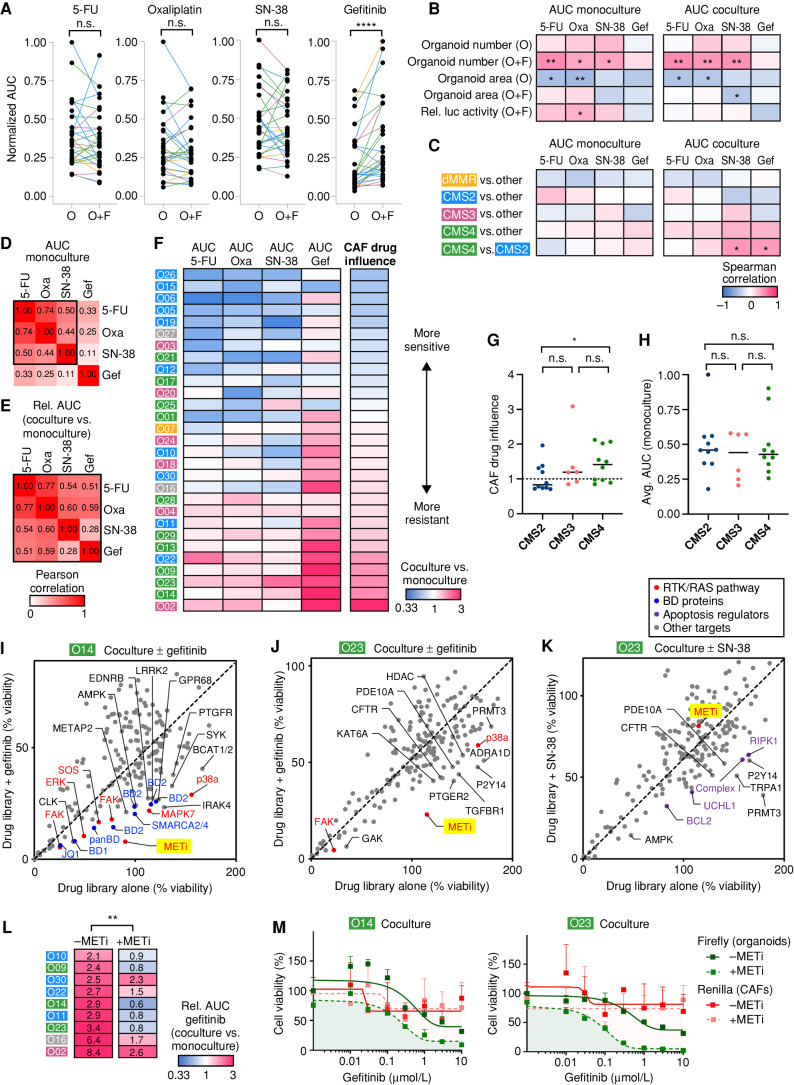
Figure 6.
Coculture exposes subtype-specific therapy resistance and individualized drug vulnerabilities. A, Pharmacotyping of four clinical drugs in biobank (n = 29, color indicates original CMS). Tumor cell viability was assessed in monocultures (O) and cocultures (O+F) by luciferase measurement. Normalized AUC was calculated by dividing the maximum AUC value for each drug. ****, P < 0.0001; n.s.,P > 0.05 (Wilcoxon matched-pairs signed rank test). B and C, Spearman correlation between drug responses and growth characteristics (B; data from Fig. 4) or original tumor subtypes (C). Organoids with >1,000 somatic alterations were defined as dMMR. Significant changes are labeled: *, P < 0.05; **, P < 0.01 (Mann–Whitney U test). Gef, gefitinib; Oxa, oxaliplatin; Rel luc, relative luciferase. D and E, Pearson correlation between different drug treatments. AUC in monoculture (D) and the relative change of drug sensitivity in the presence of CAFs (E; AUCco/AUCmono) are shown. F, Heat map of differential drug sensitivity in the presence of CAFs (AUCco/AUCmono). Data are sorted according to the CAF drug influence, representing the average relative change of all four treatments. CMS of original tumors are labeled. G and H, Subtype comparison between CAF drug influence, (G) and the average (Avg.) AUC in monoculture (H). Medians are marked. Coculture induces significantly higher resistance in CMS4- compared with CMS2-derived models. Mann–Whitney U test (*, P = 0.029; n.s., P > 0.05). I–K, Pharmacologic screens show patient- and treatment-specific resistance mechanisms. A chemogenomic library containing 186 drugs was tested in O14 and O23 in coculture with F14. Stroma-induced resistance was analyzed by comparison of the library alone or in combination with a sublethal concentration of gefitinib (I/J) or SN-38 (K). Tumor cell viability was assessed by luciferase measurement in transgenic organoids. Mean data from two experimental replicates are shown. Top hits comprise MET inhibitor (METi; BAY-474) and other RTK/RAS pathway–associated proteins (red), BD protein inhibitors (blue), and apoptosis regulators (violet). L, MET inhibitor treatment to overcome gefitinib resistance. Heat map shows differential gefitinib response in resistant cocultures (AUCco/AUCmono). Addition of 1 μmol/L BAY-474 restores sensitivity in eight of nine tested cocultures. **, P < 0.01 (Wilcoxon matched-pairs signed rank test). M, Dual luciferase assay in cocultures (O14 and O23); 1 μmol/L BAY-474 induces vulnerability of tumor cells (Firefly, green) but not of CAFs (Renilla, red). Mean viability (+SD in triplicate wells) relative to DMSO alone. Experiments were repeated twice independently. See Supplementary Fig. S11 and Supplementary Tables S9 and S10.