Table 2. Chitosan Chain Conformation Data.a.
| chitosanl | 6.49 (6.39, 6.53, 0.05) | 0.499 (0.516, 0.454, 0.024) | 24.0* | |||
| chitosant | 5.16 (3.69, 6.24, 1.01) | 0.365 (0.226, 0.486, 0.094) | 9.5 (9.9) | |||
| chitosanh | 3.32 (1.56, 4.98, 1.41) | 0.199 (0.055, 0.324, 0.109) | 1.6 (2.8) |
 ,
, 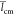 , and
, and 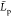 mean gyration radius, mean monomer unit
contour length, and mean chain persistence length, respectively. Bold
data: value averaged over the 10 replica (for the persistence length,
the mean value is computed from averaging the replica Ck functions and superscript * indicates the mean end to
end distance, see main text).Values in parentheses: smallest, largest,
and standard deviation data within the replica set, at the exception
of
mean gyration radius, mean monomer unit
contour length, and mean chain persistence length, respectively. Bold
data: value averaged over the 10 replica (for the persistence length,
the mean value is computed from averaging the replica Ck functions and superscript * indicates the mean end to
end distance, see main text).Values in parentheses: smallest, largest,
and standard deviation data within the replica set, at the exception
of 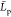 data, for which the values in parentheses
are computed from eq 15. All data in nm.
data, for which the values in parentheses
are computed from eq 15. All data in nm.
