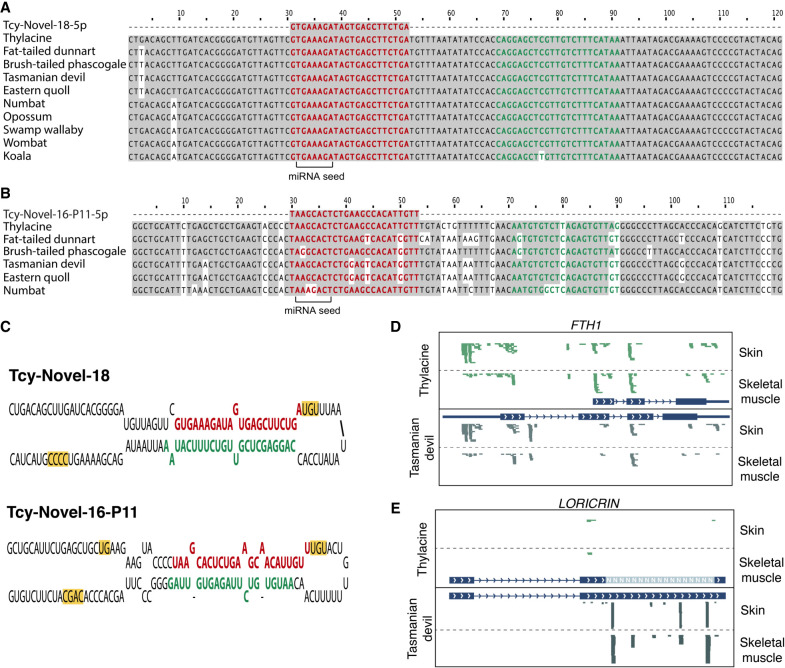
Figure 6.
Novel thylacine miRNAs and improved gene annotations guided by historical RNA sequences. Multiple sequence alignment for Tcy-Novel-18 (A) and Tcy-Novel-16-P11 (B) selected novel miRNA candidates predicted using miRDeep2 software. The 5p arm showing transcriptional evidence in the thylacine RNA sequencing data from skeletal muscle and skin tissue is highlighted in bold red. The 3p arm is highlighted in bold green. Nucleotides that are shared with respect to the thylacine species for each novel miRNA candidate are shown in gray. (C) Predicted secondary structure folding of the pri-miRNA precursor sequences (±30 nt from the pre-miRNA) for Tcy-Novel-18 and Tcy-Novel-16-P11 novel miRNA candidates. The 5p and 3p mature arms of the miRNA hairpins are highlighted in bold red and green, respectively. Processing motifs characteristic of true miRNA loci are highlighted in yellow. Examples of missing exonic annotations in the thylacine assembly are shown for transcriptional profiles (in green) obtained for FTH1 (D) and LORICRIN (E) genes, using the thylacine genome assembly as a reference. RNA sequencing data of skeletal muscle and skin thylacine tissues were aligned to the Tasmanian devil assembly (in gray) for comparison.