Fig. 1.
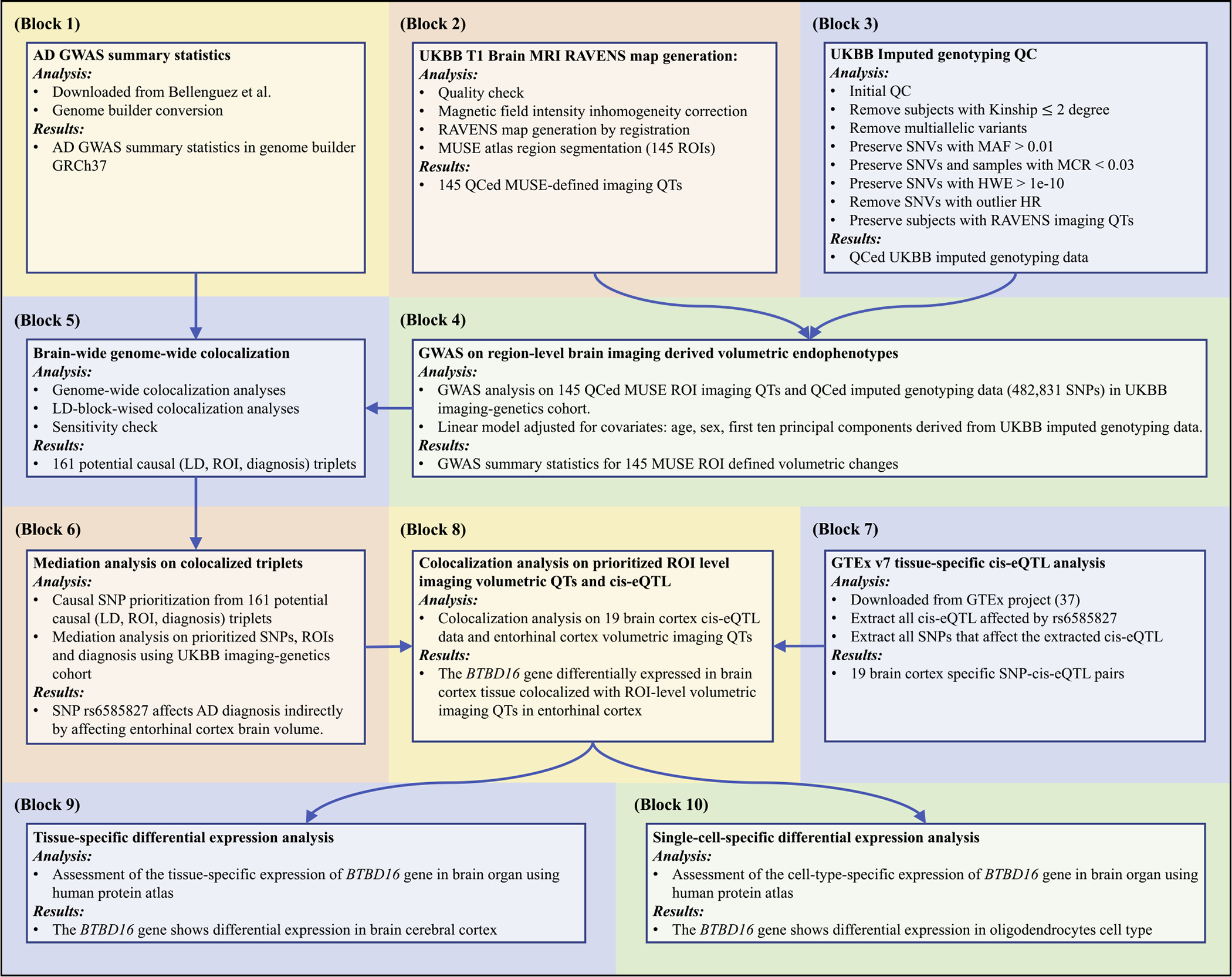
The brian-wide genome-wide colocalization framework. Block 1) AD GWAS summary statistics. Block 2) UKBB T1-weighted MRI preprocessing and RAVENS map generation using MUSE atlas (Materials and methods: UKBB imaging genomics data preprocessing). Block 3) UKBB imputed genotyping QC (Materials and methods: UKBB imaging genomics data preprocessing). Block 4) GWAS on 145 MUSE ROIs using imputed genotyping data (Materials and methods: GWAS of brain imaging quantitative traits). Block 5) LD-block-wised brain-volumetric-QTs-diagnosis colocalization analyses (Materials and methods: Brain-wide genome-wide colocalization analysis). Block 6) Mediation analyses on prioritized SNP-ROI pairs (Materials and methods: Mediation analysis). Block 7) GTEx v7 tissue-specific cis-eQTL analysis (Materials and methods: Colocalization on mediation analysis prioritized SNP-ROI pairs). Block 8) Colocalization analysis on prioritized ROIs and cis-eQTL on potential causal SNPs (Materials and methods: Colocalization on mediation analysis prioritized SNP-ROI pairs). Block 9) Tissue-specific differential expression analysis (Materials and methods: Tissue-specific differential expression analysis and single-cell-specific differential expression analysis). Block 10) Single-cell-specific differential expression analysis (Materials and methods: Tissue-specific differential expression analysis and single-cell-specific differential expression analysis). Abbreviations: AD: Alzheimer’s disease; GWAS: genome-wide association study; GRCh37: Genome Reference Consortium Human Build 37; UKBB: UK biobank; MRI: magnetic resonance imaging; RAVENS: voxel-wise regional volumetric maps; MUSE: multi-atlas region segmentation; ROI: region of interest; QC: quality control; QT: quantitative trait; SNV: single nucleotide variation; MAF: minor allele frequency; MCR: missing call rate; HWE: Hardy-Weinberg equilibrium; HR: heterozygosity rate; LD: linkage disequilibrium; SNP: single nucleotide polymorphism; cis-eQTL: cis-expression-quantitative-trait-loci; GTEx: Genotype-Tissue Expression project.
