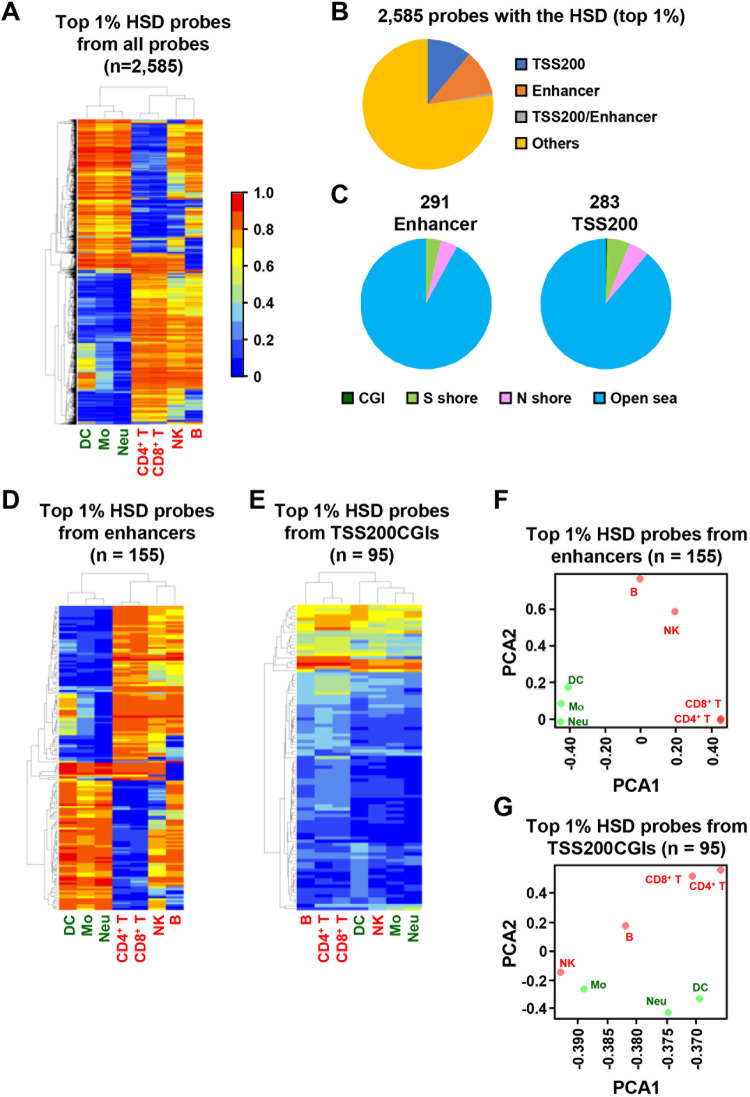
Fig 1. The presence of DNA methylation profiles specific to individual types of mouse leukocytes.
(A) Unsupervised hierarchical clustering analysis using the top 1% (2,588 probes) of the total 258,525 probes that had the HSD. The three myeloid lineage cell types and four lymphoid lineage cell types were clustered separately. (B) Origins of the 2,588 probes against gene structure. 291 (11.3%) and 283 (10.9%) of probes were derived from enhancers and TSS200, respectively. (C) Origins of the 291 enhancer and 283 TSS200 probes against CGI. The vast majority of the HSD probes were in open sea, and 0 enhancer probe and 1 TSS200 probe were in CGI. (D) Unsupervised hierarchical clustering analysis using the top 1% (155 probes) from enhancers. The three myeloid lineages and the four lymphoid lineages were classified, similar to the use of the HSD probes from all genomic regions. (E) Unsupervised hierarchical clustering analysis using the top 1% (95 probes) from TSS200CGIs. Separation of the two lineages was not clear. (F) and (G) Principal component analysis (PCA) using the HSD probes from enhancers (F) and those from TSS200 (G). Separation of the two lineages was clearer using probes from enhancers.