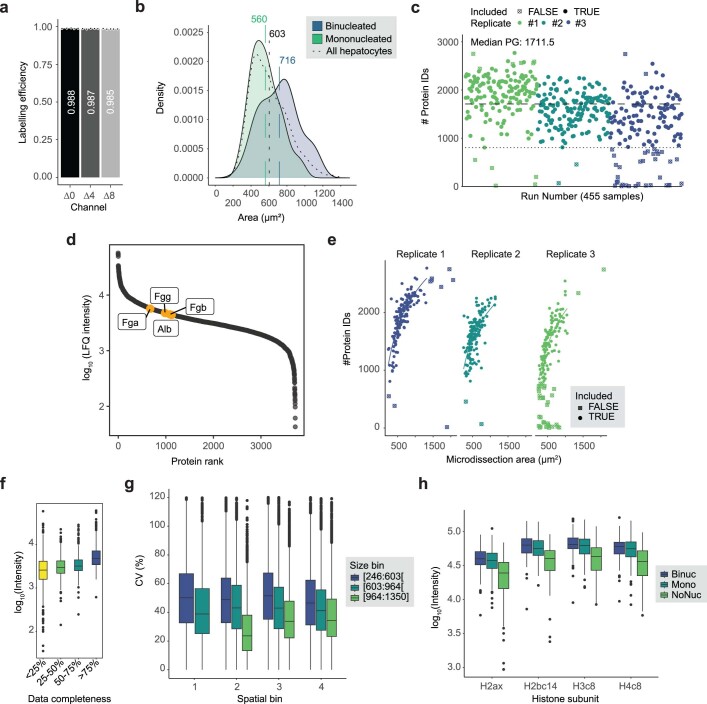
Extended Data Fig. 3. Performance overview of single-shape proteomes.
a, Labeling efficiency of 10 ng mouse liver peptide samples. Mean efficiency by intensity is stated in the bar (n = 5, mean and individual measurements). b, Density distribution of shape areas across all measured and included hepatocytes, split by visually distinguished mono- (N = 191) and binucleated (N = 99) hepatocytes. Vertical lines and numbers above are mean sizes in the respective group. c, Number of proteins per sample (N = 455). The dotted line is the median, the fine pricked line is the sample exclusion cutoff of median minus 1.5 standard deviations. Samples were measured from left to right. Shape type indicates whether the samples was included for the final analysis. d, Levels of plasma proteins in the scDVP dataset. Hba, Hbb and Hbd were not detected. e, Association between the area of the cut shape, and number of proteins. Line is a log10 regression curve. Symbols indicate whether sample was included or discarded for analysis, for exclusion criteria see Methods section. f, Percentage of proteins quantified, binned into four groups, versus log10 transformed median intensities in the respective bin. Data completeness is defined as percentage of samples across all samples in which a particular protein was quantified in. g, Coefficient of variation (CV) in bins of similarly sized shapes (color coded), and spatial bins with similar distance ratio to portal and central vein, that is similar zonation profile. h, Levels of four histone proteins shown in Fig. 2b by number of nuclei in the isolated shapes. Binuc: binucleated (N = 99); Mono: mononucleated (N = 191); NoNuc: no nucleus (N = 101). Boxes are first and third quartile, the thick line is median, whiskers are ± 1.5 interquartile range, and outliers are indicated as individual points.