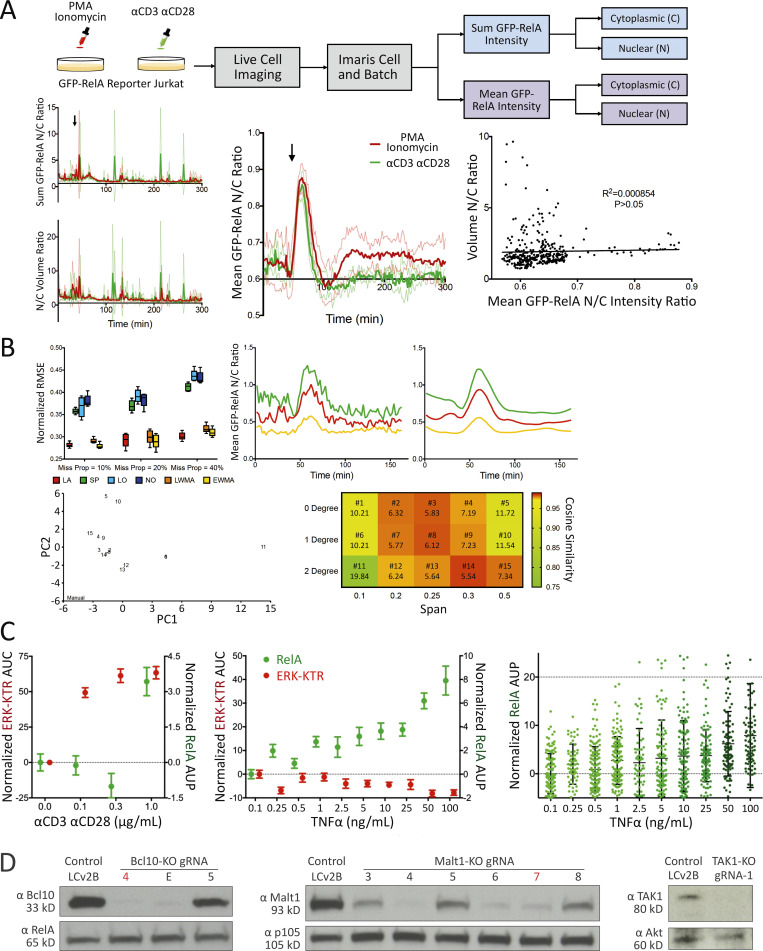
Figure S1.
TranslocQ image analysis pipelines, αCD3/αCD28 or TNFα titration data, and protein expression analysis for BCL10, MALT1, and TAK1. (A) Mean GFP nuclear to cytoplasmic intensity ratio recapitulates RelA nuclear translocation dynamics. Population response to PMA/ionomycin (red) or αCD3/αCD28 (green) stimulation calculated from GFP-RelA reporter Jurkat single-cell N/C ratio of sum GFP intensity (top left), mean GFP intensity (middle), or N/C volume ratio. Shown are mean ± s.d. of the population response at three imaging positions for each stimulation added at the indicated time point (arrow). Correlation between volume N/C ratio and GFP-RelA mean intensity N/C ratio (right). Statistical significance and coefficient of determination were calculated using a Pearson correlation test (P > 0.05). (B) Missing value interpolation and the smoothing function enable the execution of peak detection. Comparison of missing value interpolation methods at different missing data proportions using the dataset that contains 200 complete single-cell traces (top left). Shown are mean (±s.d.) of normalized root mean square error (n = 5). Representative traces for active single cells (top right) before and after smoothing with LOESS (degree = 1, span = 0.25). PCA (bottom left) of single-cell classification performed with manual observation and TranslocQ. For each method, the number of cells classified into each category (active, semi-active, inactive) at each position were considered (n = 3). Quantification accuracy of TranslocQ with different parameter sets (bottom right), represented by the cosine similarity with manual measurement. Euclidean distance to the manual data point in the PCA table is shown along with the parameter set number. (C) ERK-KTR versus NFκB-RelA dose responses to αCD3/αCD28 or TNFα. Shown are the AUP (mean ± standard error of the mean [s.e.m.]) of single-cell N/C GFP-RelA (in green) or the AUC (mean ± s.e.m.) of single-cell C/N ERK-KTR-mScarlet (in red) traces. Dose responses of RelA to TNFα titration are also shown as single-cell AUP distribution, along with the corresponding mean ± s.d. error bar. Results are representative of three technical replicates. (D) CRISPR/Cas9-mediated KO efficiency of BCL10, MALT1, and TAK1 assessed by Western blot analysis. The gRNA selected for use in Fig. 2B is highlighted for BCL10-KO (gRNA-4 in red) or MALT1-KO (gRNA-7 in red). Source data are available for this figure: SourceData FS1.