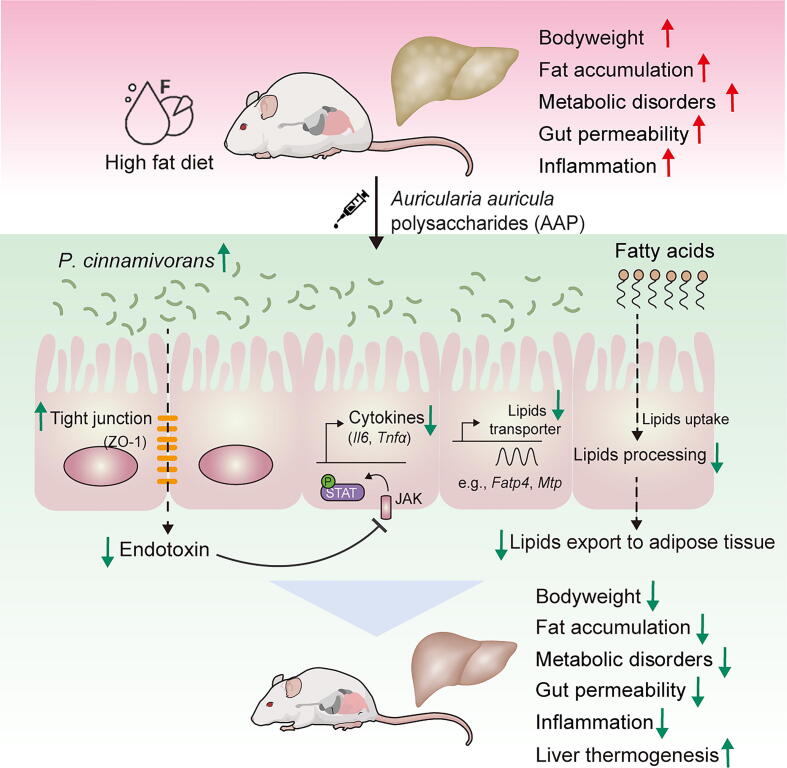
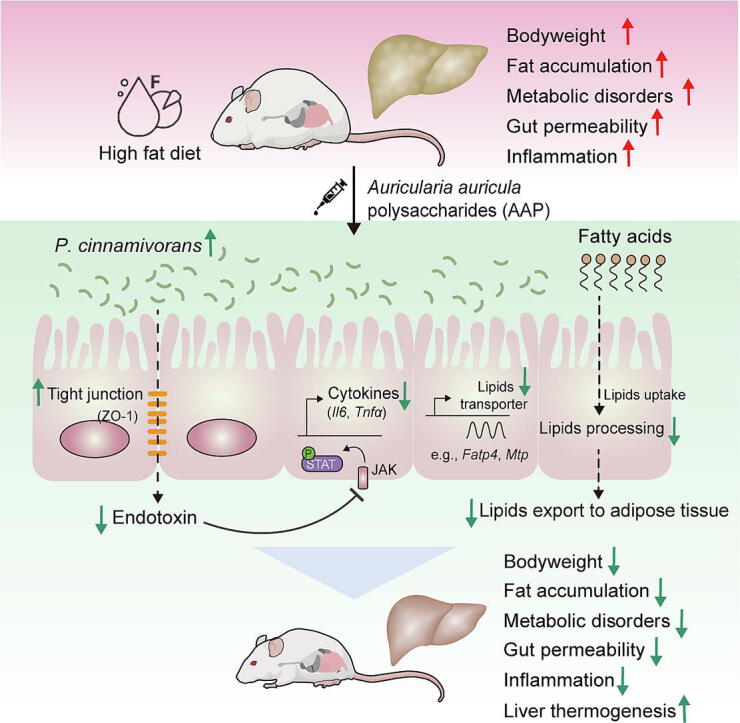
Graphical abstract
Keywords: Auricularia auricula, Polysaccharides, Obesity, Gut microbiota, Papillibacter cinnamivorans
Highlights
-
•
Auricularia auricula polysaccharides (AAP) attenuate high fat diet (HFD) induced obesity and metabolic disorders.
-
•
Anti-obesogenic effects of AAP were dependent on the gut microbiota.
-
•
Gut commensal Papillibacter cinnamivorans was a key factor for AAP reducing HFD-induced obesity.
-
•
Papillibacter cinnamivorans reduced HFD-induced obesity by regulating intestinal lipid absorption and hepatic thermogenesis.
Abstract
Introduction
Auricularia auricula is a well-known traditional edible and medical fungus with high nutritional and pharmacological values, as well as metabolic and immunoregulatory properties. Nondigestible fermentable polysaccharides are identified as primary bioactive constituents of Auricularia auricula extracts. However, the exact mechanisms underlying the effects of Auricularia auricula polysaccharides (AAP) on obesity and related metabolic endpoints, including the role of the gut microbiota, remain insufficiently understood.
Methods
The effects of AAP on obesity were assessed within high-fat diet (HFD)-based mice through obesity trait analysis and metabolomic profiling. To determine the mechanistic role of the gut microbiota in observed anti-obesogenic effects AAP, faecal microbiota transplantation (FMT) and pseudo-germ-free mice model treated with antibiotics were also applied, together with 16S rRNA genomic-derived taxonomic profiling.
Results
High-fat diet (HFD) murine exposure to AAP thwarted weight gains, reduced fat depositing and enhanced glucose tolerance, together with upregulating thermogenesis proteomic biomarkers within adipose tissue. Serum metabolome indicated these effects were associated with changes in fatty acid metabolism. Intestine-dwelling microbial population assessments discovered that AAP selectively enhanced Papillibacter cinnamivorans, a commensal bacterium with reduced presence in HFD mice. Notably, HFD mice treated with oral formulations of P. cinnamivorans attenuated obesity, which was linked to decreased intestinal lipid transportation and hepatic thermogenesis. Mechanistically, it was demonstrated that P. cinnamivorans regulated intestinal lipids metabolism and liver thermogenesis by reducing the proinflammatory response and gut permeability in a JAK-STAT signaling-related manner.
Conclusion
Datasets from the present study show that AAP thwarted dietary-driven obesity and metabolism-based disorders by regulating intestinal lipid transportation, a mechanism that is dependent on the gut commensal P. cinnamivorans. These results indicated AAP and P. cinnamivorans as newly identified pre- and probiotics that could serve as novel therapeutics against obesity.
Introduction
Obesity can be defined as a major, life-threatening chronic disease that provides detriment to the health and quality-of-life of our modern society [1]. Accumulating evidence suggests that obesity is associated with a dysregulated metabolic and immune system, which underlies the development of numerous obesity-associated comorbidities, including type II diabetes, non-alcoholic fatty liver disease, cardiovascular disease, and select cancers [2]. Halting obesity by regulating its underlying immunometabolism is therefore essential to attain better societal health and well-being. Thus, an urgent need exists for the identification of novel nutritional and therapeutic strategies for the treatment and prevention of obesity and its associated comorbidities.
Residing within the gastrointestinal tract of humans and other animals is a complex microbial community consisting of trillions of microorganisms, collectively known as the gut microbiota [3]. The compositional and functional homeostasis of these microbial populations is widely recognized to play a role in promoting both human health and the development of chronic diseases [4], including the pathogenesis of obesity and its associated comorbidities [5], [6]. Noteworthy, a double-blind, randomized controlled human study revealed that oral-fecal microbiota transplantations (FMT) combined with daily low-fermentable fiber supplementation improved insulin sensitivity in patients with severe obesity and metabolic syndrome, demonstrating the role of the gut microbiota in the development of obesity-associated metabolic disorders [7]. Dietary strategies that maintain the homeostasis of intestine-dwelling microbial populations have also been proposed as effective but least invasive methods for the prevention and treatment of obesity [8], [9]. However, additional investigations are required to gather sufficient knowledge on the intricate interplays within the trifecta of dietary intake, obesity and intestinal microbiota.
Nowadays, the interaction between natural products, especially bioactive compounds from drug-food homologous plants and fungus, and intestine-dwelling microbial populations have been widely studied [10]. Emerging evidence highlighted several bioactive constituents of naturally-occurring compounds such as non-digestible polysaccharides that could diminish obesity by regulating the composition and functions of individual intestinal microbiota [11]. For example, several animal studies have shown that mushroom polysaccharides and their dietary fibers diminish bodyweight gains and reduce insulin resistance by increasing the abundance of bespoke anti-obesogenic intestinal microbes, together with generating microbiome-derived metabolites [12], [13]. Another study identified that Sarcodon aspratus polysaccharides enhanced obesity-driven metabolic disorders and regulated intestinal dysbiosis in high-fat diet (HFD)-induced obese mice [14]. Polysaccharides derived from Hirsutella sinensis were found to exhibit anti-obesogenic and anti-diabetic effects by modulating the composition of the gut microbiota, particularly enriching the gut commensal bacterium Parabacteroides goldsteinii [15]. Arguably, due to their effects on the gut microbiota and host metabolism, the administration of natural, fungus-derived polysaccharides holds promise for the treatment of obesity.
Auricularia auricula, a well-known traditional Chinese medicinal herb, is the third most important cultivate, edible fungi worldwide, and has attracted increasing research interest for its abundance and myriad of bioactive components and health roles [16]. The fruit bodies of Auricularia auricula are rich in polysaccharides and have been historically used both as a food and as a drug in oriental countries for over 1000 years [17]. The most functional components of Auricularia auricula are considered to be the polysaccharides, which have been suggested to play important biological roles, such as antioxidant, hypoglycemic, hypolipidemic, immunomodulatory, antitumor and antiviral activities [18], [19], [20], [21], [22], [23], [24]. However, whether Auricularia auricula polysaccharides (AAP) provide direct influence over bodyweight and obesity-linked metabolic disorders remains largely unknown.
Findings of the present study reveal that polysaccharides isolated from Auricularia auricula reduced bodyweight and associated metabolic disorders in HFD-induced obese mice. Further experiments demonstrated that the gut commensal Papillibacter cinnamivorans was required for the anti-obesogenic effects of AAP. Mechanistically, it was demonstrated that P. cinnamivorans regulated intestinal lipid metabolism and liver thermogenesis by reducing the proinflammatory response and gut permeability in a JAK-STAT signaling-related manner. Collectively, this study discovered a novel gut commensal microbial species that directly influences fungal polysaccharide-based anti-obesogenic functions and further demonstrated that AAP can serve as a potential health supplement for the prevention of obesity and related comorbidities.
Materials and methods
Ethics statement
All experiments involving mice were performed in accordance with the ethical policies and procedures approved by the Animal Care Welfare Committee of Zhejiang University (No. 22703).
Isolation, purification and characterization of AAP
Auricularia auricula (Origforest, Xi’an, China) was identified and utilized as recorded in advance. AAP fractions were prepared via hot water extraction and ethanol precipitation as described previously [25]. A combination of papain enzymolysis and Sevag methods were used for deproteinization [26]. Briefly, papain was added to the hot water extract of Auricularia auricula, which worked in the pH range from 6.0 to 7.0, and the reaction was executed at 55 °C for 2 h in a water bath. To inactivate the protease, the temperature was adjusted to 100 °C and boiled for 10 min after the reaction [26]. After dialysis and freeze-drying, AAP was obtained.
The structural features of the obtained AAP were confirmed as previously described [26], [27]. The content of polysaccharides in AAP was 97.6%, which was quantified by the phenol–sulfuric acid method. The molecular weight was determined by high-performance gel filtration chromatography, which indicated that AAP was a homogeneous polysaccharide with an average molecular weight of 1065.2 kDa. High-performance liquid chromatography was employed to analyze the monosaccharide composition of AAP with pre-column derivatization of 1-phenyl-3-methyl-5-pyrazolone (PMP), it revealed that AAP was mainly composed of mannose (Man), glucose (Glc), xylose (Xyl), galactose (Gal), glucuronic acid (GlcUA) and fucose (Fuc) with molar ratios of 50.84%, 21.61%, 9.24%, 8.58%, 5.78% and 3.96%, respectively. Fourier transform infrared spectroscopy of AAP indicated the characteristic absorption peaks of the carbohydrate (Fig. S1).
Animal experimental designs
Experiment 1: AAP supplementation
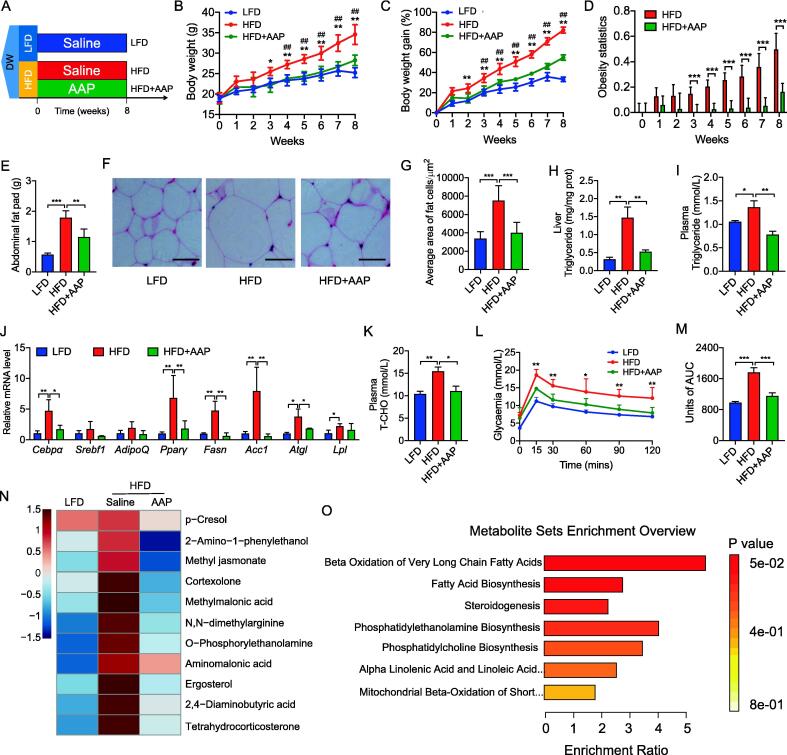
To evaluate the effect of AAP treatment on HFD-induced obesity, 30 C57BL/6 male mice aged 6 weeks were procured from Xi’an Jiaotong University. The mice were randomly divided into 3 groups (n = 10) and kept in cages (5/cage) in a controlled, specific-pathogen-free room (21℃ ± 2℃, 12 h dark-light cycle) with food and water ad libitum. Once the animals were acclimatized for one week, the diets were changed to either low-fat diet (LFD) (Diets D12450-H, Fanbo, Shanghai, China) or HFD (Diets D12451, Fanbo, Shanghai, China) (see Table S1 for the LFD and HFD formulations). As shown in Fig. 1A, mice further received a daily, 8-week intragastric gavage of either 100 μL sterile 0.9% saline with AAP (200 mg/kg bodyweight) (HFD-treated, n = 10) or 100 μL sterile 0.9% saline alone (HFD-treated, n = 10; LFD-treated, n = 10). At the pre-determined time, animals were fasted for 12 h, anesthetized, and then whole blood was obtained through retro-orbital sampling. After the mice were sacrificed, adipose tissue and organs were obtained and weighed. Fecal material was carefully collected and placed in cryotubes. The collected samples were then flash-frozen in liquid nitrogen and kept frozen at −80 °C until further analysis.
Fig. 1.
Oral administration of AAP circumvented dietary-driven obesity and metabolic disorders. (A) Mice were maintained on either a low-fat diet (LFD) or a high-fat diet (HFD) and exposed to daily AAP dosing for 8 weeks, n = 10 per treatment group. (B-D) Bodyweight (B), bodyweight gain (C), and bodyweight changes relative to the LFD reflected as obesity statistics (D). (E) Abdominal fat pad weight. (F) H&E-staining abdominal adipose tissue imaging. Scale-bar, 50 μm. (G) Adipocyte size in abdominal adipose tissues was determined from five microscopy fields for each murine using Adiposoft (Image J). (H) Hepatic triglyceride content. (I) Circulating triglycerides levels. (J) qPCR analysis of adipogenesis and lipogenesis mRNA expression in abdominal fat. (K) Circulating T-CHO levels. (L, M) OGTT and the corresponding area under the curve (AUC). (N) Heatmap of metabolites with significant variation according to the serum metabolome of HFD mice with and without AAP treatment, and of LFD mice. (O) Summary of metabolite set enrichment pathway analyses. Datasets reflected mean ± SEM. DW, distilled water. Line graphs were analyzed by two-way ANOVA, and histograms were analyzed by ordinary one-way ANOVA followed by Tukey’s post hoc test. # significant difference between LFD and HFD groups; * significant difference between HFD and HFD + AAP groups, * or #, P < 0.05, ** or ##, P < 0.01.
Experiment 2: Antibiotics treatment
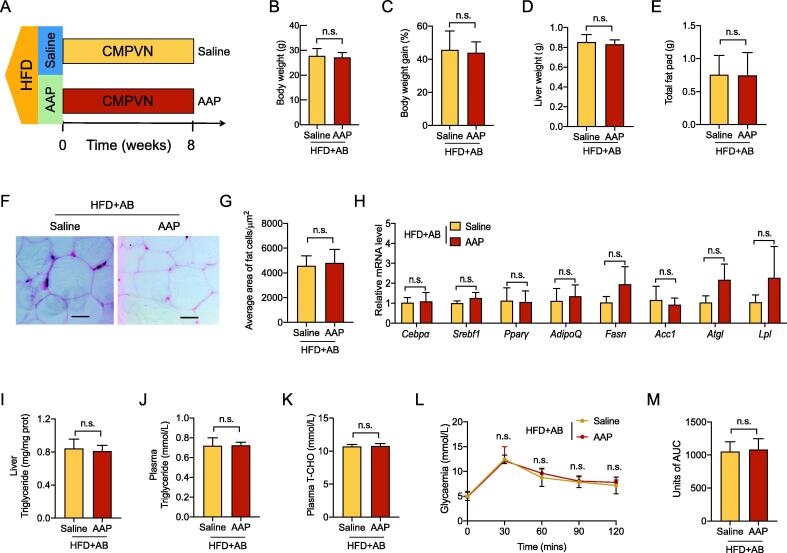
To ascertain whether anti-obesogenic effects of AAP were linked to the gut microbiota, an in vivo antibiotic treatment was performed. Briefly, as shown in Fig. 2A, 20 C57BL/6 male mice aged 6 weeks were randomly divided into 2 groups (n = 10, 5/cage) and reared in the same environment and conditions specified above. After one week of acclimatization, mice were fed an HFD and treated for 8 weeks with either AAP or 0.9% saline and sterile distilled drinking water with an antibiotic cocktail that included 50 μg/mL clindamycin (Sigma, USA), 50 μg/mL metronidazole (Sigma, USA), 50 μg/mL penicillin (Sigma, USA), 25 μg/mL vancomycin (Sigma, USA), and 50 μg/mL neomycin (Sigma, USA). The blood and tissue sample collection and processing methods used were similar to those described in Experiment 1.
Fig. 2.
The intestinal microbiota was crucial for AAP against obesity. (A) HFD mice were exposed to an antibiotic combination of clindamycin, metronidazole, penicillin, vancomycin and neomycin (CMPVN) and either saline or AAP throughout all 8 weeks, n = 10 per group. (B-C) Bodyweight (B), bodyweight gain (C). (D) Liver weight. (E) Abdominal fat pad weight. (F) H&E-staining abdominal adipose tissue imaging. Scale-bar, 50 μm. (G) Adipocyte size in abdominal adipose tissues was determined from five microscopy fields for each murine using Adiposoft (Image J). (H) qPCR analysis of adipogenesis and lipogenesis mRNA expression within abdominal fat, expressed relative to the housekeeping mRNA, Gapdh. (I) Hepatic triglyceride content. (J-K) Circulating triglycerides and T-CHO levels. (L, M) OGTT and the corresponding AUC. Datasets reflected mean ± SEM. DW, distilled water. Line graphs were analyzed by two-way ANOVA, and histograms were analyzed by t-test. ** P < 0.01.
Experiment 3: Fecal microbiota transplantations (FMT)
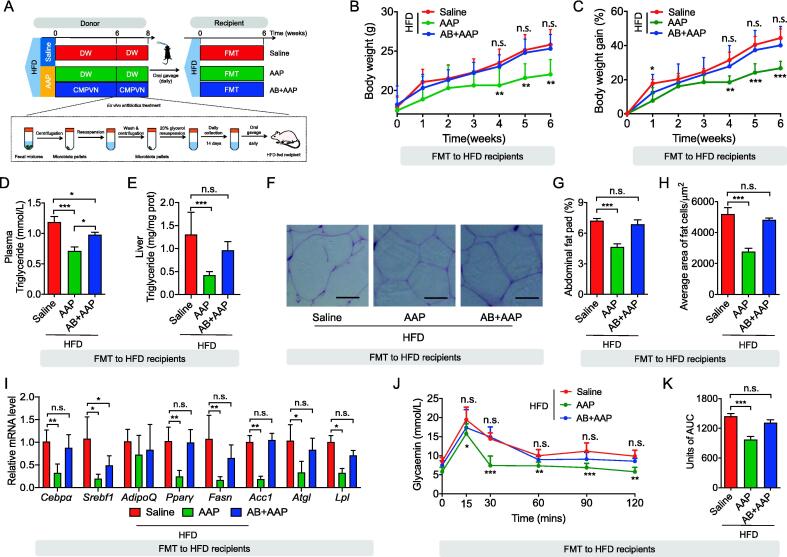
To further ascertain the role of the gut microbiota in the anti-obesogenic effects of AAP, FMT was performed using fecal material obtained from HFD-fed donor mice from experiments 1 and 2 that were treated for 8 weeks with either 0.9% saline, AAP or AAP plus antibiotics. From weeks 6 to 8, fecal samples from each donor mouse were prepared in anaerobiosis and counted by light microscopy through methylene blue staining, and then frozen at −80 °C with 10% glycerol until use.
Fecal samples were thawed and prepared for the FMT as previously described [28]. In brief, we pooled fecal material from different mice in the same group. 80–110 mg of collected feces were homogenized in 1 ml of sterile saline in autoclaved tubes. After centrifugation at 2,000 g at 4 °C for 1 min, the supernatants were transferred to new tubes and centrifuged for 5 min at 15,000 g to precipitate the microbiome. 30 C57BL/6 male mice aged 5 weeks were randomly divided into 3 groups (n = 10, 5/cage) and reared in the same environment and conditions specified above. After one week of acclimatization, as shown in Fig. 3A, all mice were fed HFD and treated daily with 100 μL of fecal microbiota transplants from each donor group via oral gavage for 6 weeks. The blood and tissue sample collection and processing methods used were similar to those described in Experiment 1.
Fig. 3.
FMT from AAP-exposed mice diminished obesity and metabolic disorders within HFD mice. (A) Fecal microbiota from HFD mice given saline or AAP with/without antibiotics were transplanted into HFD recipients, n = 10 per group. (B-C) Bodyweight (B), bodyweight gain (C). (D) Hepatic triglyceride content. (E) Circulating triglycerides level. (F) H&E-staining abdominal adipose tissue imaging. Scale-bar, 50 μm. (G) Abdominal fat pad weight. (H) Adipocyte dimensions in abdominal adipose tissue were determined through five microscopy fields for each murine using Adiposoft (Image J). (I) qPCR analysis of adipogenesis and lipogenesis mRNA expression in abdominal fat, expressed relative to the housekeeping mRNA, Gapdh. (J, K) OGTT and corresponding AUC. Datasets reflected mean ± SEM. DW, distilled water. Line graphs were analyzed by two-way ANOVA, and histograms were analyzed by ordinary one-way ANOVA followed by Tukey’s post hoc test. * P < 0.05, ** P < 0.01 and *** P < 0.001.
Experiment 4: Papillibacter cinnamivorans treatment
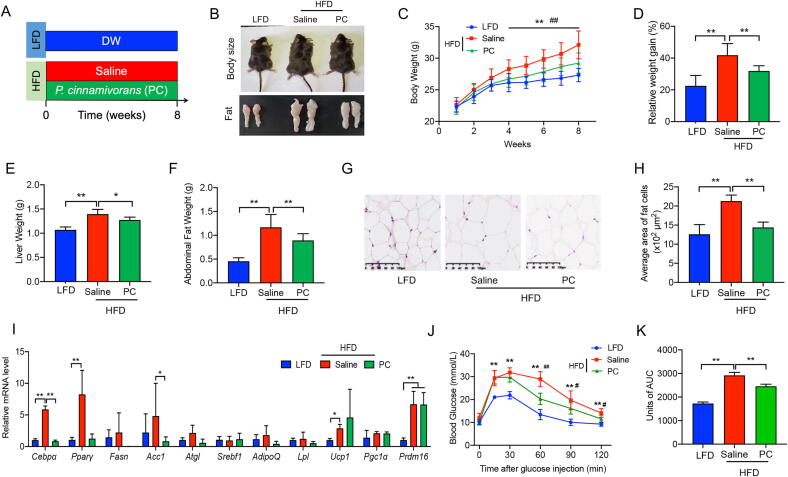
P. cinnamivorans (ATCC 700879) was purchased from the American Type Culture Collection. The bacterium was grown at 37 °C in a Whitley DG250 anaerobic chamber (Don Whitley, UK) with mixed anaerobic gas (20% carbon dioxide, 80% nitrogen). Anaerobicity was confirmed using an anaerobic indicator (Oxoid, UK). P. cinnamivorans was cultivated on anaerobic Cinnamate Medium (ATCC Medium 1788). The live bacteria were harvested during the logarithmic growth phase and counted by light microscopy through methylene blue staining. After centrifugation, the bacteria were aliquoted as stock at 4 × 108 colony-forming units/ml and stored at −80 °C until use. 30 C57BL/6 male mice aged 5 weeks were randomly divided into 3 groups (n = 10, 5/cage) and reared in the same environment and conditions specified above. After one week of acclimatization, as shown in Fig. 6A, mice were fed an LFD/HFD and treated for 8 weeks with either 4 × 107 colony-forming units of P. cinnamivorans in 100 μL by oral gavage or 0.9% saline. The blood and tissue sample collection and processing methods used were similar to those described in Experiment 1.
Fig. 6.
P. cinnamivorans treatment protected mice against dietary-driven obesity and metabolic disorders. (A) LFD-fed mice and high-fat diet (HFD)-fed mice were treated daily with saline or P. cinnamivorans (PC) by oral gavage for 8 weeks. (B) Representative picture of body size, and fat pad. (C-D) Bodyweight (C), bodyweight gain (D). (E) Liver weight. (F) Abdominal fat pad weight. (G) H&E-staining abdominal adipose tissue imaging. Scale-bar, 100 μm. (H) Adipocyte dimensions and quantity within abdominal adipose tissue were determined from five microscopy fields for each murine. (I) qPCR analysis of Cebp and Pparγ mRNA expression in abdominal fat, expressed relative to the housekeeping mRNA, Gapdh. (J, K) OGTT and the corresponding AUC. Datasets reflected mean ± SEM. DW, distilled water. Line graphs were analyzed by two-way ANOVA, and histograms were analyzed by ordinary one-way ANOVA followed by Tukey’s post hoc test. # significant difference between LFD and HFD groups; * significant difference between HFD and HFD + PC groups, * or #, P < 0.05, ** or ##, P < 0.01.
Bodyweight measurements
The bodyweight of each mouse was assessed by using a digital scale. In terms of bodyweight change, the bodyweight of each mouse was measured on days 1, 3 and 5 of each week. The ratio between treatment bodyweight and initial bodyweight was then calculated, as well as the change in bodyweight relative to the LFD.
Measurement of lipid markers levels, lipopolysaccharides (LPS) and cytokines
Triglycerides, total cholesterol (T-CHO), low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), and LPS were assessed by commercial colorimetric kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. Colonic and serum concentrations of TNF-α and IL-6 were determined using commercial enzyme-linked immunoassay kits (eBioscience, San Diego, USA) according to the manufacturer’s instructions.
Oral glucose tolerance test (OGTT)
Mice were fasted for 12 h prior to the OGTT while proving sterile drinking water ad libitum. Each mouse then received an intragastric gavage of a 50% glucose solution (2 g/kg bodyweight; Aladdin, Shanghai, China) with blood glucose levels further measured by glucometer (Yuwell, Jiangsu, China) at 0, 15, 30, 60, 90, and 120 min post- gavage.
Morphology analysis
The fresh tissue from the abdominal fat pads was fixed in 4% paraformaldehyde, dehydrated, transparentized, paraffin-coated, and consequently segmented and stained through hematoxylin and eosin (H&E) as previously reported [29]. Six mice were randomly selected from each group and then a random section of adipose tissue from each mouse was captured using a Leica DM3000 Microsystem, with the size of adipocytes determined through Image J v1.53 (https://imagej.nih.gov/ij/) at 10x magnification.
Immunofluorescence analysis
Paraformaldehyde-fixed and paraffin-embedded sections of the colon were used for immunofluorescence as previously described [30]. Briefly, the sections were incubated with primary antibodies (1:200 dilution) specific for ZO-1, Occludin and Claudin-1. Then, fluorescently labeled antibodies were used as secondary antibodies (1:50 dilution) and examined with a Leica fluorescence microscope (Keyence, Osaka, Japan). The nuclei of the colonic epithelium were stained with 4′,6-diamidino-2-phenylindole.
Transmission electron microscopy (TEM)
Colonic tight junction structures and hepatic fat deposits were observed by TEM as previously described [30]. Ultrathin sections were obtained using a diamond knife and stained with uranyl acetate and lead citrate before examination by TEM (JEM-1011; JEOL USA, Peabody, MA). Digital electron micrographs were acquired with a CCD camera system (AMT Corp., Denver, MA).
Western blot analysis
Tissues were lysed on ice in an SDS-PAGE sample buffer. Proteins were transferred onto a polyvinylidene fluoride membrane (MilliporeSigma, Billerica, MA), then were blotted and quantified as described previously [31]. The antibodies, including ZO-1, Occludin, and Claudin-1, were purchased from Proteintech (Wuhan, China) [25].
RNA extraction and quantitative real-time PCR (qPCR) analysis
Total RNA of the jejunum and adipose tissues was isolated and preserved using TRIzol (Invitrogen, USA) as previously described [32]. Previously validated RT-qPCR primers for genes of interest (Table S2) were then used for the relative quantification of the gene expression through SYBR Green Master Mix (Roche, Germany) on the StepOne Plus RT-PCR System (Applied Biosystems, USA) by using 2–ΔΔCT methodology.
Serum metabolomic profiling by GC-TOF/MS
Sample preparation and metabolite determination by GC-TOF/MS were carried out according to our previous report [33], [34]. Briefly, after ultrasound treatment and centrifugation, the supernatant was dehydrated, derivatized and mixed with 30 μL methoxy-amination hydrochloride (20 mg/mL in pyridine), 40 μL of bis (trimethylsilyl) trifluoroacetamide regent (BSTFA, 1% TMCS, v/v) and 5 μL fatty acid methyl esters (in chloroform), respectively. GC-TOF/MS was performed using an Agilent 7890 gas chromatography platform (Agilent Technologies, USA), combined with a Pegasus HT time-of-flight mass spectrometer (LECO Chroma TOF PEGASUS HT, LECO, USA). The MetaboAnalyst 5.0 platform (https://www.metaboanalyst.ca/home.xhtml) was employed for the bioinformatic analysis. Briefly, sample normalization (normalization by sum), data transformation (log-transformed) and data scaling (auto-scaling) were carried out for data normalization. Pattern recognition multivariate analyses, such as principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA), were also performed. The compounds with variable importance of projection (VIP) > 1.0 in PLS-DA and P-value < 0.05 in one-way ANOVA followed by Tukey’s post hoc test were identified as significantly different metabolites between groups. The enrichment analysis was further performed using the significantly different metabolites based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
RNA sequencing and analysis
Library preparation of RNA sequencing and high-throughput sequencing was performed by Novogene (Beijing, China). Briefly, total RNA extracted from murine hepatic and colonic tissues were used for library preparation. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. More than 44,020,824 reads for each sample were obtained. Raw data were filtered, corrected, and mapped to locust genome sequence via the HISAT software. Gene expression levels were measured using the criteria of reads per kb per million mapped reads. DEGs were detected using the DESeq software [35]. The P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 were assigned as differentially expressed. KEGG enrichment was performed by the Clusterfile software. Significance analysis was performed by Fisher’s Exact Test.
Gut microbiota and bioinformatic analysis
Sequencing of 16S rRNA gene amplicons and the analysis of obtained data was performed as previously described [36]. Feces from 6 mice were randomly selected from experiments 1 to 3. Total bacterial DNA was extracted using the Solarbio fecal genomic DNA extraction kit (Beijing, China) in combination with mechanical cell lysis by bead beating as previously reported [37]. Characteristics for the extraction were evaluated through 1% agar-gel electrophoresis. PCRs were conducted targeting the V3-V4 region with universal primers, 338F: 5′-ACTCCTACGGGAGGCAGCA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′, or the V4 region with universal primers, 515F: 5ʹ-GTGCCAGCMGCCGCGG-3ʹ and 806R: 5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ. Regarding further extraction, purification, and quantification of PCR products, the QuantiFluor-ST platform (Promega, USA) and AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, China) were utilized, or the Thermo Scientific GeneJET Gel Extraction and DNA Cleanup Micro Kit (Thermo Fisher, USA) were employed following the manufacturer’s protocols. The sequencing libraries were constructed using the TruSeqTM DNA Sample Prep Kit (SanDiego, CA, USA) or Plus Fragment Library Kit (Thermo Fisher, USA), and sequenced on the Illumina MiSeq PE300 platform by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) or IonS5TMXL platform (Thermo Fisher, USA) by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China), respectively.
The microbial community analysis was carried out according to our previous report [37]. Briefly, read processing and quality control were conducted using the QIIME 1 pipeline. Filtered sequences were clustered into operational taxonomic units (OTUs) using the UPARSE pipeline. As the number of OTUs detected is a factor of sequencing depth, 16S rRNA gene amplicon data from each experiment were first normalized to the minimum number of sequences. Then, the relevant characterizing sequences were taxonomically categorized utilizing the Ribosomal Database Project (RDP) MultiClassifier tool [38] and the Greengenes database [39]. α-Diversity indices such as Chao1 were calculated. Ordination plots for β-diversity metrics were generated by principal coordinates analysis (PCoA) based on Bray Curtis, unweighted UniFrac and weighted UniFrac distances [40]. The ANOSIM test was performed to determine whether the separation of groups was significant. Differentially abundant taxa were identified by the linear discriminant analysis (LDA) effect size (LEfSe) analysis [41]. Furthermore, differences between the two groups in taxonomic and diversity data were compared with Welch’s two-sided t-test. For multigroup comparison, one-way analyses of variance (ANOVA) in combination with Kruskal-Wallis test were applied. We indicated and confirmed the taxonomy of significantly different OTUs using NCBI blastn, EzBioCloud, and Ribosomal Database Project Seqmatch [42], [43]. Correlations between phenotypes, significantly different metabolites and microbes were computed with the Pearson test in R using the corplot package (R Core Team, 2014).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 6.01). All datasets reflected mean ± standard error of the mean (SEM). Line graphs were analyzed by two-way ANOVA, histograms of more than two groups were analyzed by ordinary one-way ANOVA followed by Tukey’s post hoc test, and t-tests were used on two groups of histograms. For the phenotypic data, we checked the normality and homoscedasticity (variance equivalence) for ANOVA, while for microbiota abundance we used the Kruskal-Wallis test to compare differences among groups. Statistical significance was considered at P < 0.05.
Results
AAP attenuated HFD-induced obesity and metabolic disorders
To investigate the effect of AAP on diet-induced obesity, mice were fed an HFD for 8 weeks augmented through daily oral administration of AAP or saline (negative control) (Fig. 1A). Following these 8 weeks, bodyweight for the HFD group increased significantly (Fig. 1B). However, AAP was shown to attenuate dietary-driven weight gains from week-2 onwards by 38.88% (Fig. 1B-D). The histological analysis corroborated that AAP administration decreased abdominal fat deposits and adipocyte size (Fig. 1E-G), which was accompanied by lower liver and heart weights, but kidney and spleen weights showed no differences (Fig. S2A-D). Further analysis of the abdominal fat showed that adipogenesis (for Pparγ and Cebpα) and lipogenesis genes (for Acc1 and Fasn) were upregulated by HFD, while downregulated by AAP treatment (Fig. 1J). In addition, HFD-induced hepatic steatosis and dyslipidemia were inhibited by AAP, as demonstrated by reduced triglyceride accumulation within the liver (Fig. 1H) and reduced levels of plasma triglyceride (Fig. 1I), cholesterol (T-CHO, Fig. 1K), and low-density lipoprotein (LDL) (Fig. S2F). However, no changes in plasma HDL were observed (Fig. S2E). Furthermore, we found that the HFD-induced hyperglycemia was attenuated by AAP as shown by lower plasma glucose levels during an OGTT (Fig. 1L-M), suggesting AAP enhanced the clearance of glucose post-consumption.
To further elucidate the effects of AAP on HFD-induced obesity and associated metabolic disorders, metabolomic analyses were performed to compare the serum metabolome of mice only fed an HFD with those also treated with AAP, with mice fed LFD as the control. Unsupervised PCA of the 197 detected metabolites revealed a clear separation of the serum metabolomes among HFD, HFD + AAP, and LFD-fed mice (Fig. S3A). The data suggest that AAP treatment benefited the HFD-induced metabolic disorder by affecting serum metabolite profiles, which was also supported by the PLS-DA analysis (Fig. S3B). Further analysis of individual metabolites suggested that the output of 11 metabolites elevated by HFD was completely reversed to normal levels by AAP (Fig. 1N), which included 2-amino-1-phenylethanol, methyl jasmonate, cortexolone, methylmalonic acid, ergosterol, tetrahydrocorticosterone. Correlation analyses showed that these metabolites were positively linked to obesity phenotypes, especially bodyweight (Fig. S3C). Metabolic pathway analysis demonstrated that the significantly different metabolites were associated with metabolic pathways involved in fatty acid metabolism (Fig. 1O). Together, these results demonstrated that the oral administration of AAP prevented HFD-induced obesity and its metabolic disorders, likely by relating fatty acid metabolism.
Anti-obesogenic effects of AAP were dependent on the gut microbiota
To determine whether the gut microbiota played a role in AAP-induced effects on obesity and related disorders, HFD mice were exposed to an antibiotic cocktail (Fig. 2A). As shown in Fig. 2B-C, AAP failed to attenuate the HFD-induced bodyweight gain when mice were treated with antibiotics, which was further supported by similar organ weights (Fig. 2D, Fig. S4A-C). The fat of mice treated with or without antibiotics also showed no differences in abdominal fat weight, adipocyte size, or expression of adipogenesis-related genes (Fig. 2E-H). Furtherly, antibiotic treatment abolished the effects of AAP on HFD-induced metabolic disorders, including liver triglyceride levels, peripheral triglyceride, T-CHO, and LDL levels (Fig. 2I-K, Fig. S4D-E), as well as glucose clearance (Fig. 2L-M). Together, the anti-obesogenic influences of AAP were dependent on the presence of a diverse gut microbial community.
Fig. 4.
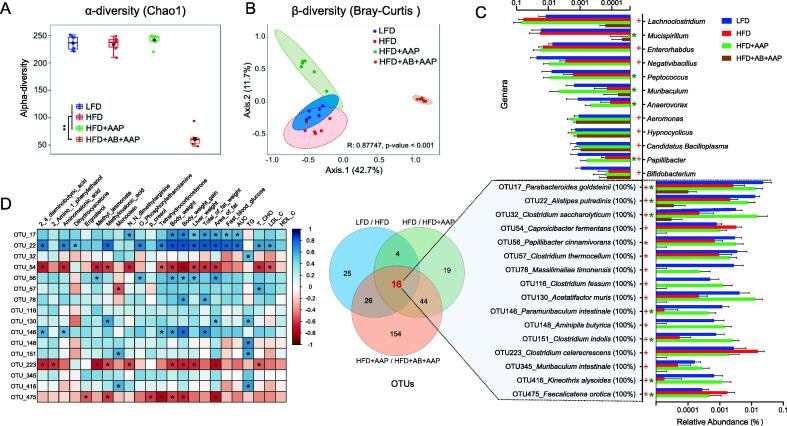
Alternation of intestinal microbiota in obese mice treated with AAP. (A) α-diversity assessed through Chao1 index. (B) β-diversity analysis by PCoA on Bray-Curtis distance. (C) Differentially abundant genera and operational taxonomic units (OTUs) from 16S rRNA genomic-derived fecal microbiota assessment using LEfSe analysis. (D) Correlation analysis for all 16 recognized bacterial species and obesity-related endpoints. False-discovery-rate corrections for multiple analyses were employed. + significant difference between LFD and HFD groups, * significant difference between HFD and HFD + AAP groups, + or *, P < 0.05.
FMT confirmed the role of gut microbiota in the anti-obesogenic effects of AAP
To further elucidate the role of the gut microbiota in AAP-induced effects, FMT was applied to conventional mice consuming an HFD. Fecal microbiota from HFD mice treated with either AAP without antibiotics, AAP with antibiotics, or saline (control) were transplanted into HFD recipient mice (Fig. 3A). Compared to the control mice, FMT from AAP-treated mice markedly lowered bodyweight gains and organ weights (Fig. 3B-C, Fig. S5A-C). However, FMT recipients from AAP plus antibiotics-treated mice were indistinguishable from HFD mice (Fig. 3B-C, Fig. S5A-C). Similarly, when compared to HFD-FMT recipients, AAP-FMT recipients showed significantly lower levels of triglyceride accumulation within the liver and peripheral circulation (Fig. 3D-E) together with serum T-CHO, HDL and LDL (Fig. S5D-F), which suggests that transplantation of the fecal microbial community modified by AAP also reduced lipid metabolism. As further validation, abdominal fat deposition and expression levels for adipogenesis-related genes were analyzed. While no differences were detected between the mice receiving FMT from the controls or the AAP treatment with antibiotics, a significant reduction was detected for the mice receiving AAP-FMT (Fig. 3F-I). Regarding the OGTT, AAP-FMT recipients also had a markedly reduced rise in blood glucose levels with the antibiotics treatment abolishing the effects of AAP-FMT (Fig. 3J-K). Overall, the findings further confirmed the role of microbiota in the anti-obesogenic effects of AAP.
Fig. 5.
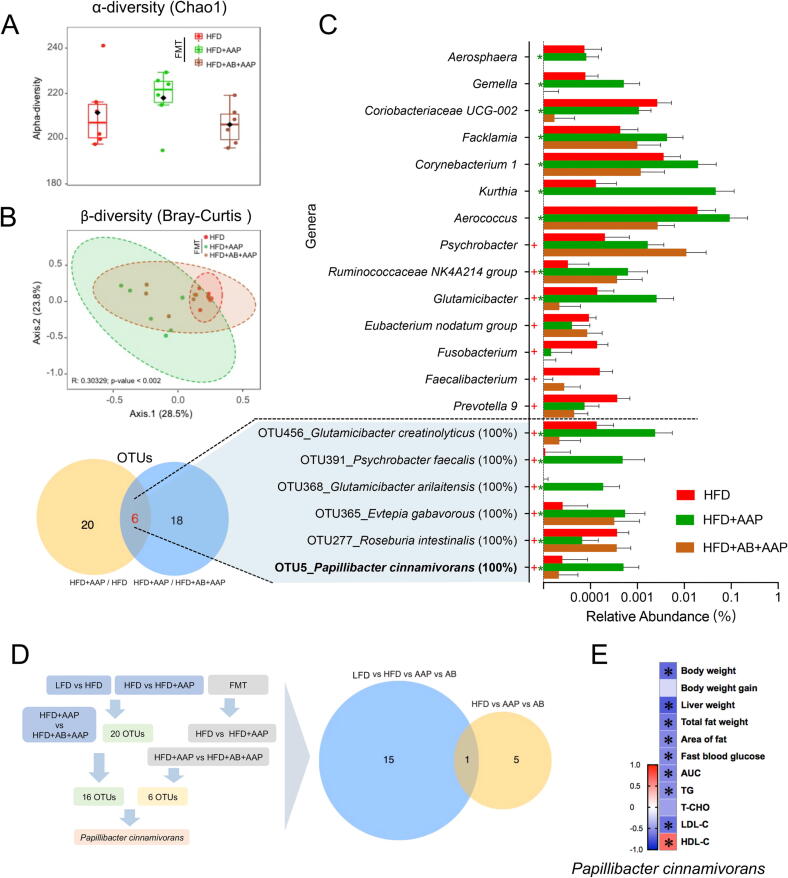
P. cinnamivorans was identified as the key bacteria for AAP reducing diet-induced obesity. (A) α-diversity assessed using the Chao1 index. (B) β-diversity analysis by PCoA on Bray-Curtis distance. (C) Differentially abundant genera and operational taxonomic units (OTUs) from 16S rRNA genomic-derived fecal microbiota analysis using LEfSe analysis. (D) Analytical strategy for key bacteria identification. (E) Correlation analysis for P. cinnamivorans and obesity trends. False-discovery-rate corrections for multiple testing were employed. + significant difference between LFD and HFD groups, * significant difference between HFD and HFD + AAP groups, + or *, P < 0.05.
AAP treatment shifted the gut microbiota of HFD-induced obese mice
To further understand the role of gut microbiota in the alleviation of obesity by AAP, the intestine-dwelling microbiome makeup was investigated through 16S rRNA gene amplicon sequencing. While α-diversity showed no differences between groups, inter-group β-Diversity, as visualized by PCoA, showed four separate clusters dependent on the treatment, suggesting differences in gut microbiota composition in response to the HFD, AAP, and antibiotic interventions (Fig. 4 A-B, Fig. S6A-B).
LEfSe analysis was employed to identify bacterial taxa that shifted by AAP in HFD mice. At the genus level, compared to LFD, there were eight genera shown to be changed by HFD (five genera increased and three genera decreased), including Lachnoclostridium, Enterorhabdus, Negativibacillus, Aeromonas, Hypnocyclicus, Candidatus Bacilloplasma, Papillibacter, and Bifidobacterium (Fig. 4C). Compared with the HFD mice, AAP treatment significantly decreased the level of Mucispirillum, and increased levels of Peptococcus, Muribaculum, Anaerovorax, and Papillibacter, which were eliminated by antibiotics (Fig. 4C). Notably, only levels of the genus Papillibacter were inhibited by HFD and consequently recovered by AAP (Fig. 4C).
OTUs showing a significant difference between AAP and the other HFD groups were searched against the GenBank sequence database. Globally, 71 bacterial OTUs were markedly shifted through HFD, while in comparison to HFD, AAP shifted 83 bacterial OTUs (Fig. 4C). This study further evaluated OTU overlap between the comparison of three different experimental classes, including the normal diet versus HFD groups, HFD versus HFD + AAP groups, and HFD + AAP versus HFD + AB + AAP groups. And 16 OTUs were obtained (Table S3), including P. cinnamivorans, Parabacteroides goldsteinii, Alistipes putredinis, Clostridium saccharolyticum, Caprobacter fermentans, Clostridium thermocellum, Massilimaliae timonensis, Clostridium fessum, Acetatifactor muris, Paramuribaculum intestinale, Aminipila butyrica, Clostridium indolis, Clostridium celerecrescens, Muribaculum intestinale, Kineothrix alysoides, and Faecalicatena orotica (Fig. 4C). Consequently, this investigation focused on the possibility of obesity-linked trends being associated with intestinal levels of those bacterial species regulated through AAP. The correlation analysis demonstrated that bacterial levels, including OTU17 (P. goldsteinii), OTU22 (A. putredinis) and OTU56 (P. cinnamivorans), were significantly and negatively correlated to obesity (Fig. 4D). These results demonstrated that AAP modulated HFD-induced obesity in a gut microbiota-specific manner.
P. cinnamivorans was a key factor for AAP reducing HFD-induced obesity
Since the anti-obesity effects of AAP were transferable through FMT, the impact of transferring donor fecal microbiota on microbial diversity was evaluated, with 16S rRNA genomic sequencing conducted across recipient fecal samples. Results included a spectrum of microbiota-shifting influences from the pooled screening, predominantly not affecting microbiota alpha diversity (Fig. 5 A). To account for the effects of treatment on β-diversity, this investigation found significant differences among HFD, AAP, and antibiotics cohorts, which demonstrated that FMT reproduced the effects of AAP on the gut microbiota of HFD-induced obese mice (Fig. 5B, Fig. S6C-D). A total of 54 significantly different bacterial genera were identified, of which 14 genera were common among the three groups. Among these genera, compared with AAP-FMT recipients, 9 genera were significantly altered by antibiotic-treated fecal colonies, including Aerosphaera, Gemella, Coriobacteriaceae, Facklamia, Corynebacterium, Kurthia, Aerococcus, Ruminococcaceae, Glutamicibacter, and 7 genera were regulated in the mice transplanted with fecal microbiota from HFD mice, including Psychrobacter, Ruminococcaceae, Glutamicibacter, Eubacterium nodatum group, Fusobacterium, Faecalibacterium, Prevotella (Fig. 5C). Furthermore, using the GenBank sequence database, the differing OTUs among the three groups were analyzed. There were 26 OTUs identified as shifted species in AAP-FMT treated mice versus HFD-FMT treated mice, and 24 OTUs were altered in the antibiotic-FMT treated mice compared with AAP-FMT treated mice (Fig. 5C). This study focused on the six-OTU-overlap in the two communities (Table S4) and found that five of them were increased in the AAP-FMT cohort, though not in the antibiotic-FMT cohort, in comparison with the HFD-FMT cohort, besides the OTU277_Roseburia intestinalis (Fig. 5C). Obesity-reducing occurred in mice treated with AAP and AAP-FMT and therefore, the functional gut-commensal bacteria against obesity should exist in the fecal samples for both cohorts. Surprisingly, only OTU5_P. cinnamivorans was identified from the comparison between these two sequencing assays (Fig. 5D). The results of correlation analysis showed that the relative abundance of P. cinnamivorans had a negative correlation to obesity trends (Fig. 5E). These data indicated that P. cinnamivorans was potentially the key factor for AAP in reducing HFD-induced obesity.
P. cinnamivorans treatment protected mice against HFD-induced obesity and metabolic disorders
To confirm that P. cinnamivorans promoted anti-obesogenic effects in HFD mice, a commercialized strain of P. cinnamivorans or saline (control) was administered daily for 8 weeks within HFD mice (Fig. 6A). After 4 weeks of treatment with P. cinnamivorans, the relative abundance of P. cinnamivorans in fecal samples significantly increased (Fig. S6E). Furthermore, bodyweight gains were attenuated relative to HFD mice, and these effects persisted throughout the final 4 weeks of the study (Fig. 6B-D). Treatment with P. cinnamivorans was also shown to effectively reduce HFD-induced obesity traits, such as liver and abdominal fat weight (Fig. 6E-F), and these results were accompanied by the mean sizes of adipocytes and average area of fat cells from abdominal adipose tissue (Fig. 6G-H). We then assessed the expression of fat metabolism related genes in the abdominal fat by comparing control and P. cinnamivorans treated mice. We found P. cinnamivorans also significantly decreased the expression of Cebpα and Pparγ (Fig. 6I). Moreover, to further characterize the metabolic phenotype, an OGTT was performed. The results showed that blood glucose concentrations were reduced at 60, 90, and 120 min after glucose injection in mice treated with P. cinnamivorans, in comparison to HFD mice (Fig. 6J-K). These data suggested that P. cinnamivorans treatment could protect mice against HFD-induced obesity and metabolic disorders.
P. cinnamivorans alleviated the inflammatory response and enhanced intestinal barrier function in HFD-induced obesity mice
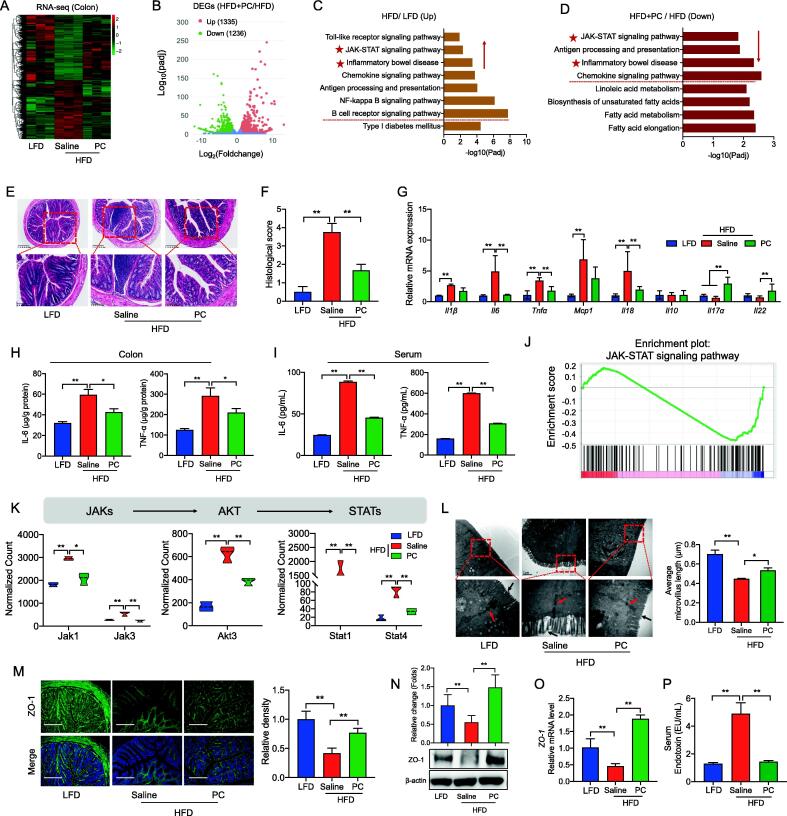
Having established that P. cinnamivorans reduced obesity phenotypes, we next wondered about the mechanisms underlying these effects. To this end, we performed a bulk RNA sequencing (RNA-seq) analysis to more comprehensively analyze gene expression profiles on colon tissues of the LFD mice, HFD-fed mice, and the HFD-P. cinnamivorans-treated mice (Fig. 7A, and Fig. S7A). Differential expression analysis identified 1479 differential expression genes (DEG) for the HFD mice compared with that of the LFD mice, consisting of 818 upregulated and 661 downregulated genes (Fig. S7B). Meanwhile, of the 2571 genes obtained in P. cinnamivorans treatment compared with that in the HFD group, 1335 were upregulated and 1236 were downregulated (Fig. 7B). KEGG enrichment analysis indicated that inflammation-related pathways positioned in the overlap of the two clusters of HFD_up versus LFD and PC_down versus HFD, including the JAK-STAT signaling pathway, antigen processing and presentation, inflammatory bowel diseases, and chemokine signaling pathway (Fig. 7C-D). Therefore, to further investigate the role of P. cinnamivorans on intestinal inflammation mediated by HFD, we measured the intestinal inflammatory response. Morphological analysis by H&E staining revealed that HFD induced considerable tissue injury with extensive ulceration of the epithelial layer, edema, crypt damage to the bowel wall and infiltration of granulocytes and mononuclear cells into the mucosa (Fig. 7E-F). In contrast, P. cinnamivorans treatment reduced the histological evidence of HFD-induced colon inflammation (Fig. 7E-F). To characterize the effects of P. cinnamivorans against HFD-induced inflammation, the expression of inflammatory markers was quantified. As shown in Fig. 7G, P. cinnamivorans treatment significantly reduced the expression of proinflammatory cytokines, such as Tnfα, Il6, and Il18. To validate these results, the secretion of TNF-α and IL-6 in the colon and serum was analyzed, which was consistent with the results of gene expression (Fig. 7H-I). Furtherly, to gain more insights into the molecular mechanisms of the anti-inflammatory effects of P. cinnamivorans, we performed a gene-set enrichment analysis (GSEA) of the JAK-STAT signaling pathway (Fig. 7J). The data showed that P. cinnamivorans treatment markedly inhibited the activation of JAK-STAT signaling in mice fed HFD, through analysis of the transcription level of Jak1, Jak3, Akt3, Stat1 and Stat4 (Fig. 7K). These suggested that P. cinnamivorans treatment suppressed the HFD-induced intestinal inflammatory response via the JAK-STAT signaling-related pathway.
Fig. 7.
P. cinnamivorans alleviated the inflammatory response and enhanced intestinal barrier function in HFD-induced obesity mice. (A) Heatmap of the differentially expressed genes (DEGs) of colon identified by RNA-seq. (B) Volcano plot showing DEGs of HFD + PC versus HFD. (C) KEGG analysis of up-regulated genes from DEGs of HFD versus LFD. (D) KEGG analysis of down-regulated genes from DEGs of HFD + PC versus HFD. (E-F) Serial sections of colon tissues were stained with H&E (E), and Histological scores (F). Scale-bar, 100 μm. (G) qPCR analysis of inflammatory marker expression in the colon, expressed relative to the housekeeping mRNA, Gapdh. (H-I) Secretion of TNF-α and IL-6 in colonic tissue (H) and serum (I) determined by ELISA. (J) Enrichment plot of JAK/STAT signaling pathway from GSEA. (K) RNA-seq expression data for JAK-STAT signaling genes. (L) The morphological changes of intestinal tight junctions were observed under transmission electron microscopy (TEM, 80,000 × magnification). (M) Immunofluorescence analysis of ZO-1 (green) and DAPI (blue). Scale-bar, 100 μm. (N) The expression of ZO-1 was assessed by Western blot. The right panel shows the relative protein levels quantified by densitometry and normalized to β-actin. (O) qPCR analysis of ZO-1 expression in the colon, expressed relative to the housekeeping mRNA, Gapdh. (P) Serum endotoxin (lipopolysaccharides, LPS) level. Datasets reflected mean ± SEM. One-way ANOVA and Tukey’s post hoc test. * P < 0.05, ** P < 0.01 and *** P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A previous study demonstrated that the attenuation of inflammation is related to intestinal permeability, which relies on tight junctions [44]. To investigate the effects of P. cinnamivorans on gut permeability, immunofluorescent staining and TEM were used to demonstrate the formation of tight junctions. TEM observation showed indistinct tight junction, reduced density and widened intercellular space in the intestinal tissues of mice fed HFD, whereas P. cinnamivorans treatment protected the intestine against HFD-induced damage (Fig. 7L). The presence of tight junctional complexes was suggested by immunofluorescent staining of the cell layers for tight junction markers, including Claudin-1, Occludin and ZO-1. Notably, we found P. cinnamivorans treatment effectively restored the expression of ZO-1, but not Claudin-1 and Occludin (Fig. 7M and Fig. S8A-B). Consistently, the protein and gene expression level of ZO-1 also supported the protective effects of P. cinnamivorans on HFD-damaged intestinal barrier function (Fig. 7N-O, and Fig. S8C). Additionally, compared with HFD mice, serum endotoxin or LPS levels were also significantly reduced in P. cinnamivorans treated mice, which indicated P. cinnamivorans treatment decreased the gut permeability of HFD mice (Fig. 7P). In summary, the anti-obesogenic and anti-inflammatory effects of P. cinnamivorans in obese animals were related to the enhancement of intestinal barrier function.
Fig. 8.
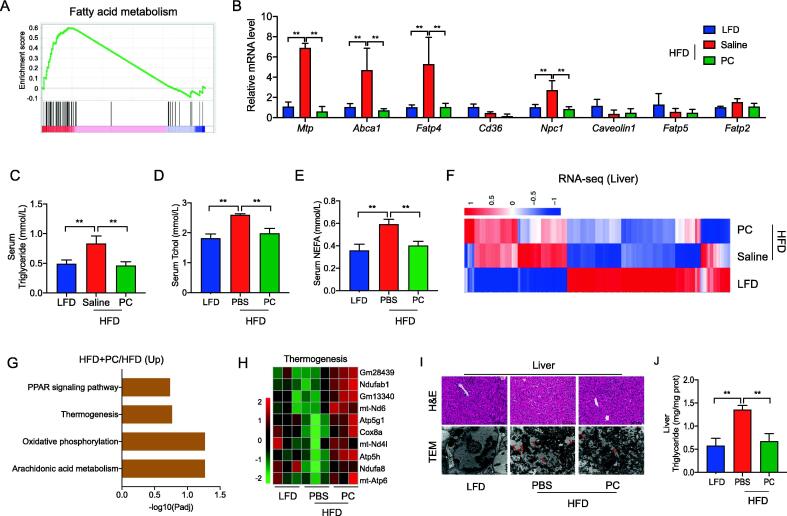
P. cinnamivorans attenuated HFD-induced obesity by regulating intestinal lipids absorption and hepatic thermogenesis. (A) Enrichment plot of fatty acid metabolism pathway from GSEA. (B) qPCR analysis of fatty acids transporters expression in the colon, expressed relative to the housekeeping mRNA, Gapdh. (C-E) Triglyceride (C), cholesterol (D), and non-esterified free fatty acid (NEFA, E) levels in serum. (F) Heatmap of the differentially expressed genes (DEGs) of liver identified by RNA-seq. (G) KEGG analysis of up-regulated genes from DEGs of HFD + PC versus HFD. (H) Heatmap of relative expression of selected thermogenesis-regulated genes from the RNA-seq dataset. (I) Liver steatosis was evaluated by H&E staining and TEM (80,000 × magnification). Scale-bar, 100 μm. (J) Hepatic triglyceride level. Datasets reflected mean ± SEM. One-way ANOVA and Tukey’s post hoc test. * P < 0.05, ** P < 0.01 and *** P < 0.001.
P. cinnamivorans reduced HFD-induced obesity by regulating intestinal lipid absorption and hepatic thermogenesis
In contrast to HFD mice, the lower expression genes in HFD and then the treatment with P. cinnamivorans were also significantly enriched in lipid metabolism-related pathways, such as fatty acid metabolism, biosynthesis of unsaturated fatty acids, fatty acid elongation, and linoleic acid metabolism (Fig. 8A, and Fig. S9A-C). These results prompted us to examine whether P. cinnamivorans mediated the anti-inflammatory and anti-obesogenic effects in HFD-fed mice by regulating intestinal lipids absorption. Using qPCR analysis, we confirmed the reduction in mRNA coding for colonic and jejunal key lipid transporters, including Mtp, Abca1, Fatp4 and Npc1, in P. cinnamivorans treated mice (Fig. 8B and Fig. S10A). This change was associated with significantly decreased serum levels of triglycerides, cholesterol and free fatty acids (Fig. 8C-E). The notion was also verified by downregulating the expression of these lipid transporters in AAP-treated mice and AAP-FMT mice, in contrast to HFD-fed mice (Fig. S10B-C).
Fig. 9.
Schematic representation of the working model. AAP selectively enhanced P. cinnamivorans, a commensal bacterium with reduced presence in HFD mice to attenuate obesity and the associated dysregulated metabolism. Mechanistically, P. cinnamivorans regulated intestinal lipids metabolism and liver thermogenesis by reducing the proinflammatory response and gut permeability in a JAK-STAT signaling-related manner.
Subsequently, we profiled the liver transcriptomes into three groups (Fig. 8F). From the results of KEGG annotation, it can be found that there were certain KEGG pathways related to obesity in PC_up versus HFD, particularly in the thermogenesis pathway (Fig. 8G). Therefore, we interrogated our data concerning the expression of thermogenesis pathway-related genes. The data show that genes related to thermogenesis signaling are upregulated in the liver of P. cinnamivorans treated mice compared to saline control (Fig. 8H). To further examine the effects of P. cinnamivorans on thermogenesis in the liver, sections of the liver were stained with H&E and TEM respectively. The results showed that P. cinnamivorans treatment promoted liver fatty degeneration to alleviate HFD-induced fat deposition (Fig. 8I). The finding was further supported by the low triglyceride levels in P. cinnamivorans treated mice (Fig. 8J). Taken together, these data further confirmed the role of P. cinnamivorans regarding anti-obesogenic effects by AAP, which attenuated obesity by regulating intestinal lipids absorption and liver thermogenesis.
Discussion
The gut microbiome has been critically linked to many aspects of metabolic syndrome, rendering it a potential target for the treatment of obesity and associated comorbidities [45]. The diet and particularly dietary carbohydrates are key determinants for the composition and activity of the intestinal microbiome. Bacterial species such as Akkermansia muciniphila [46], Faecalibacterium prausnitzii [47], and Propionibacterium freudenreichii [48] as well as non-digestible carbohydrates such as arabinoxylan [49], resistant starches [42], Ganoderma lucidum polysaccharides [50], and Hirsutella sinensis polysaccharides [15] have already shown promise for use as next-generation probiotics and prebiotics for the treatment of obesity and its dysregulated immunometabolism, respectively. In the present study, we demonstrated that routine-daily administration of AAP in HFD-fed mice could alleviate dietary-driven obesity and related metabolic disorders (Fig. 9). This research defines several essential mechanisms that underlie the anti-obesity effects of AAP, including the role of AAP-induced shifts within the gut microbial community in the observed health benefits. Remarkably, gut commensal P. cinnamivorans was identified as a key regulator in AAP treatment reversion of HFD-induced obesity and metabolic disorders. Further evaluation of plausible mechanisms suggests that enrichment of P. cinnamivorans reduced inflammatory responses and gut permeability in a JAK-STAT signaling-related manner, which regulated intestinal lipid metabolism and liver thermogenesis. Thus, our findings support the potential of using P. cinnamivorans as a probiotic supplement to ameliorate obesity-associated metabolic abnormalities.
Although previous studies have shown that A. auricula polysaccharides possessed antioxidant activity [51], modulated intestinal microbiota [52], and lowered blood glucose levels [53], the possibility that such polysaccharides regulate intestine-dwelling microbiome and diminish bodyweight in vivo was never previously studied. Therefore, we established a diet-induced obesity model to evaluate the anti-obesity effect of AAP and investigated the role of intestinal microbiota in the process. Our observations were that AAP could effectively decrease bodyweight gain, fat deposition and glucose tolerance. Polysaccharides, as a dietary fiber, have been reported to be degraded in the hindgut of animals and influenced the abundance and diversity of intestinal microbiota [54]. Intriguingly, a recent study that transferred the gut microbiota of obese individuals to HFD-fed mice conveyed larger weight gains and higher levels of obesity-associated metabolic disorders than fecal transfer from lean individuals [5]. The pseudo-germ-free mice model in our study, which applied combinatory antibiotic treatments to deplete the gut microbiota, demonstrated a loss of beneficial effects of AAP after gut microbiome deprivation, supporting the possibility that AAP may preserve gut microbiota homeostasis. The anti-obesity effects of AAP are transferrable through fecal transplantation, which also supports the concept that obesity is associated with altered gut microbiota. However, gnotobiotic models would be needed to verify the role of the gut microbiota. Our results suggest that the gut microbiota can be modulated by dietary intervention or fecal transfer and that AAP may be used as a prebiotic to induce specific gut microbiota shifts that associate with reduced weight gain, inflammation and metabolic disorders in obese individuals.
To further explore the mechanistic role of the gut microbiota in the alleviation of obesity by AAP, we measured the microbiota composition in obese mice treated with AAP. We observed that AAP supplementation in HFD-fed mice restored the gut microbiota community observed in LFD-fed mice, including the probiotic P. goldsteinii, which was previously reported to reduce obesity [15]. However, some other bacteria with negative associations with obesity, such as Akkermansia muciniphila [55] and Barnesiella spp. [56], were not detected in the present study. This observation suggests that AAP may produce anti-obesity effects by modifying the levels of other specific bacterial species, such as P. cinnamivorans, Muribaculum and Anaerovorax. Accordingly, we compared the donors' and the recipients' fecal microbiota composition and identified P. cinnamivorans as a key factor for AAP reducing diet-induced obesity, which was further confirmed in an HFD-induced obese model using a commercial strain of P. cinnamivorans. The phenotype of obesity traits and metabolic disorders showed that the anti-obesogenic effect of AAP was mainly produced by P. cinnamivorans, a strictly anaerobic, Gram-positive, non-sporulating and mesophilic bacterium that can be widely found in shea cake and animal intestines [57], [58], [59]. P. cinnamivorans is one of the species of the Papillibacter genus (family Ruminococcaceae), and the other species is an unclassified Papillibacter [60]. Moreover, a recent study indicated that it was elevated in humans on the Mediterranean diet, suggesting the beneficial function of P. cinnamivorans for humans [61]. Rensen et al., [62] analyzed the fecal microbiota constitution of 28 subjects (BMI 18.6–60.3 kg/m2) through phylogenetic microarray profiling and found that P. cinnamivorans was strongly associated with local and systematic inflammation within obesity. Similarly, a study of individual, cohort-specific intestinal microbiota profiles suggested a link between P. cinnamivorans and tissue-specific insulin sensitivity among overweight males [63]. Overall, these findings suggest that gut commensal P. cinnamivorans is a novel probiotic with the potential to be used as a therapeutic target for the attenuation of obesity and its associated comorbidities. However, a great degree of variation has been reported in the literature for the effects of probiotics on host health endpoints, which is dependent on the species/strains administered. Here we investigated the effects of P. cinnamivorans (ATCC 700879), which provides promising data to encourage future characterization of other strains within the genus. The potential anti-obesity role of other publicly available strains that are closely related to Papillibacter, such as Faecabilacterium and Subdoligranulum genera, also needs further attention and exploration.
As for the molecular mechanisms by which P. cinnamivorans reduces diet-induced obesity, our results from RNA-seq have demonstrated a key role in reducing inflammation. The reduced levels of cytokines in the colon and serum independently validated this notion. The current model of HFD-induced chronic inflammation and obesity-related disorders is explained largely by dysbiosis of the gut microbiota and increased levels of LPS in the systemic circulation [50]. Excessive fat consumption has been previously shown to increase intestinal permeability, allowing LPS to enter the entero-hepatic circulation [64]. Thus, chronically elevated circulating gut-derived LPS or “metabolic endotoxemia” could result in sustained systemic inflammation through the activation of TLR4 signaling [65]. Moreover, a previous study demonstrated that overproduction of proinflammatory cytokines such as TNF-α, IL-6 and IL-18 in obese animals may be responsible for the development of chronic inflammation and metabolic disorders [66]. Similarly, a study on water extract of Ganoderma lucidum mycelium found that it could enhance phosphorylation of AKT and decrease proinflammatory cytokine expression in the liver, leading to improved insulin sensitivity [50]. Our results indicate that P. cinnamivorans supplementation improves gut barrier integrity, reduces metabolic endotoxemia, and alleviates cytokine production in the colon and serum of HFD-fed obese mice. Furthermore, we identify the JAK-STAT signaling as the major molecular mechanism responsible for these effects. Nonetheless, we cannot rule out the possibility that P. cinnamivorans may also directly affect proinflammatory signaling pathways such as TLR2, which has been reported to control adiposity and insulin resistance [67].
The traditional perception of inflammation in the context of obesity has focused on the detrimental impact of inflammatory mediators on the pathogenesis of secondary metabolic derangements associated with obesity [68]. Our experiments demonstrate that the administration of P. cinnamivorans reduces the expression of genes involved in intestinal fatty acid absorption and enhances hepatic thermogenesis in HFD-fed mice. Manipulating thermogenesis could increase energy expenditure and improve metabolism [69]. These results suggest that P. cinnamivorans can attenuate obesity through a combination of two pathways, namely lowering lipid absorption and promoting hepatic thermogenesis. This conclusion is consistent with a previous study that showed that elevated hepatic thermogenesis could improve metabolic health and lessen weight gain [70].
In conclusion, the results of this in vivo murine investigation suggest that AAP attenuated obesity and the associated dysregulated metabolism by modulating inflammatory responses, gut permeability, and lipid transportation/metabolism in a gut microbiota-dependent manner. Such findings provide a basis for the application of AAP as a prebiotic while also demonstrating the need for further evaluation of the gut commensal P. cinnamivorans as a potential next-generation probiotic for the treatment and prevention of obesity and its metabolic comorbidities.
Compliance with ethics requirements
The Ethics Committee of Zhejiang University carefully reviewed the protocol of this study and then approved our study (Approval No. 22703), and all the tests were performed referring to the National Institutes of Health Guidelines for Animal Research (Guide for the Care and Use of Laboratory Animals, NIH Publication No.8023, revised 1996).
CRediT authorship contribution statement
Xin Zong: Investigation. Hao Zhang: Investigation, Methodology. Luoyi Zhu: Investigation. Edward C. Deehan: . Jie Fu: Methodology. Yizhen Wang: . Mingliang Jin: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32022079, 32002185, U21A20249), Zhejiang Provincial Natural Science Foundation of China (Grant Nos. LZ20C170005, LQ21C170002) and Fundamental Research Funds for the Central Universities (Grant Nos. 2020-KYY-517102-0001, 2022QZJH46).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.08.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Das M., Ellies L.G., Kumar D., Sauceda C., Oberg A., Gross E., et al. Time-restricted feeding normalizes hyperinsulinemia to inhibit breast cancer in obese postmenopausal mouse models. Nat Commun. 2021;12(1):565. doi: 10.1038/s41467-020-20743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nauck M.A., Jensen T.J., Rosenkilde C., Calanna S., Buse J.B. Investigators LPCobotLT: Neoplasms reported with liraglutide or placebo in people with type 2 diabetes: results from the leader randomized trial. Diabetes Care. 2018;41(8):1663–1671. doi: 10.2337/dc17-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong X., Fu J., Xu B.C., Wang Y.Z., Jin M.L. Interplay between gut microbiota and antimicrobial peptides. Anim Nutr. 2020;6(4):389–396. doi: 10.1016/j.aninu.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer F., Backhed F. The gut microbiota - masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 5.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome (vol 499, pg 97, 2013). Nature 2014, 506(7488). [DOI] [PubMed]
- 7.Mocanu V., Zhang Z.X., Deehan E.C., Kao D.H., Hotte N., Karmali S., et al. Fecal microbial transplantation and fiber supplementation in patients with severe obesity and metabolic syndrome: a randomized double-blind, placebo-controlled phase 2 trial. Nat Med. 2021;27(7):1272. doi: 10.1038/s41591-021-01399-2. [DOI] [PubMed] [Google Scholar]
- 8.Scott K.P., Antoine J.M., Midtvedt T., van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre M., Venema K. The art of targeting gut microbiota for tackling human obesity. Genes Nutr. 2015;10(4):472. doi: 10.1007/s12263-015-0472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu Y., He D., Li W., Wang M., Zhao S., Liu L., et al. Hepatoprotective effect of citrus aurantium l. against apap-induced liver injury by regulating liver lipid metabolism and apoptosis. Int J Biol Sci. 2020;16(5):752–765. doi: 10.7150/ijbs.40612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., Zhong M., Yu T., Chen L., Shi L., Zong J., et al. Polysaccharide extracted from WuGuChong reduces high-fat diet-induced obesity in mice by regulating the composition of intestinal microbiota. Nutr Metab (Lond) 2020;17:27. doi: 10.1186/s12986-020-00442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delannoy-Bruno O., Desai C., Raman A.S., Chen R.Y., Hibberd M.C., Cheng J.Y., et al. Lebrilla CB et al.: Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature. 2021;595(7865):91. doi: 10.1038/s41586-021-03671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma G.X., Du H.J., Hu Q.H., Yang W.J., Pei F., Xiao H. Health benefits of edible mushroom polysaccharides and associated gut microbiota regulation. Crit Rev Food Sci. 2021 doi: 10.1080/10408398.2021.1903385. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Liu J., Yan C., Zhang C., Pan W., Zhang W., et al. Sarcodon aspratus polysaccharides ameliorated obesity-induced metabolic disorders and modulated gut microbiota dysbiosis in mice fed a high-fat diet. Food Funct. 2020;11(3):2588–2602. doi: 10.1039/c9fo00963a. [DOI] [PubMed] [Google Scholar]
- 15.Wu T.R., Lin C.S., Chang C.J., Lin T.L., Martel J., Ko Y.F., et al. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut. 2019;68(2):248–262. doi: 10.1136/gutjnl-2017-315458. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y., Wang L., Zhang D., Li R., Cheng T., Zhang Y., et al. Comparative transcriptome analysis reveals relationship of three major domesticated varieties of Auricularia auricula-judae. Sci Rep. 2019;9(1):78. doi: 10.1038/s41598-018-36984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaskheli S.G., Zheng W., Sheikh S.A., Khaskheli A.A., Liu Y., Soomro A.H., et al. Characterization of Auricularia auricula polysaccharides and its antioxidant properties in fresh and pickled product. Int J Biol Macromol. 2015;81:387–395. doi: 10.1016/j.ijbiomac.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T.L., Chen J., Hu Y., Wang D., Fan Y., Wang J., et al. In vitro antiviral activity of sulfated Auricularia auricula polysaccharides. Carbohydr Polym. 2012;90(3):1254–1258. doi: 10.1016/j.carbpol.2012.06.060. [DOI] [PubMed] [Google Scholar]
- 19.Li C., Mao X., Xu B. Pulsed electric field extraction enhanced anti-coagulant effect of fungal polysaccharide from Jew's ear (Auricularia auricula) Phytochem Anal. 2013;24(1):36–40. doi: 10.1002/pca.2376. [DOI] [PubMed] [Google Scholar]
- 20.Zeng W.C., Zhang Z., Gao H., Jia L.R., Chen W.Y. Characterization of antioxidant polysaccharides from Auricularia auricular using microwave-assisted extraction. Carbohydr Polym. 2012;89(2):694–700. doi: 10.1016/j.carbpol.2012.03.078. [DOI] [PubMed] [Google Scholar]
- 21.Bao Z., Yao L., Zhang X., Lin S. Isolation, purification, characterization, and immunomodulatory effects of polysaccharide from Auricularia auricula on RAW264.7 macrophages. J Food Biochem. 2020;44(12):e13516. doi: 10.1111/jfbc.13516. [DOI] [PubMed] [Google Scholar]
- 22.Zhao R., Cheng N., Nakata P.A., Zhao L., Hu Q. Consumption of polysaccharides from Auricularia auricular modulates the intestinal microbiota in mice. Food Res Int. 2019;123:383–392. doi: 10.1016/j.foodres.2019.04.070. [DOI] [PubMed] [Google Scholar]
- 23.Kong X., Duan W., Li D., Tang X., Duan Z. Effects of polysaccharides from Auricularia auricula on the immuno-stimulatory activity and gut microbiota in immunosuppressed mice induced by cyclophosphamide. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.595700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., An X., Chen Y., Deng Y., Niu H., Ma R., et al. Effects of Auricularia auricula polysaccharides on gut microbiota and metabolic phenotype in mice. Foods. 2022;11(17) doi: 10.3390/foods11172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zong X., Cheng Y., Xiao X., Fu J., Wang F., Lu Z., et al. Protective effects of sulfated polysaccharide from Enterobacter cloacae Z0206 against DSS-induced intestinal injury via DNA methylation. Int J Biol Macromol. 2021;183:861–869. doi: 10.1016/j.ijbiomac.2021.04.182. [DOI] [PubMed] [Google Scholar]
- 26.Kai L.X., Zong X., Jiang Q., Lu Z.Q., Wang F.Q., Wang Y.Z., et al. Protective effects of polysaccharides from Atractylodes macrocephalae Koidz. against dextran sulfate sodium induced intestinal mucosal injury on mice. Int J Biol Macromol. 2022;195:142–151. doi: 10.1016/j.ijbiomac.2021.12.042. [DOI] [PubMed] [Google Scholar]
- 27.Zong X., Xiao X., Kai L.X., Cheng Y.Z., Fu J., Xu W., et al. Atractylodis macrocephalae polysaccharides protect against DSS-induced intestinal injury through a novel lncRNA ITSN1-OT1. Int J Biol Macromol. 2021;167:76–84. doi: 10.1016/j.ijbiomac.2020.11.144. [DOI] [PubMed] [Google Scholar]
- 28.Zhu L., Fu J., Xiao X., Wang F., Jin M., Fang W., et al. Faecal microbiota transplantation-mediated jejunal microbiota changes halt high-fat diet-induced obesity in mice via retarding intestinal fat absorption. Microb Biotechnol. 2022;15(1):337–352. doi: 10.1111/1751-7915.13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong X., Xiao X., Jie F., Cheng Y.Z., Jin M.L., Yin Y.L., et al. YTHDF1 promotes NLRP3 translation to induce intestinal epithelial cell inflammatory injury during endotoxic shock. Sci China Life Sci. 2021;64(11):1988–1991. doi: 10.1007/s11427-020-1909-6. [DOI] [PubMed] [Google Scholar]
- 30.Zong X., Cao X.X., Wang H., Xiao X., Wang Y.Z., Lu Z.Q. Cathelicidin-WA facilitated intestinal fatty acid absorption through enhancing PPAR-gamma dependent barrier function. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zong X., Zhao J., Wang H., Lu Z.Q., Wang F.Q., Du H.H., et al. Mettl3 deficiency sustains long-chain fatty acid absorption through suppressing Traf6-dependent inflammation response. J Immunol. 2019;202(2):567–578. doi: 10.4049/jimmunol.1801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong X., Xiao X., Shen B., Jiang Q., Wang H., Lu Z., et al. The N6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Res. 2021;49(10):5537–5552. doi: 10.1093/nar/gkab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin M., Wang J., Zhang H., Zhou H., Zhao K. Simulated weightlessness perturbs the intestinal metabolomic profile of rats. Front Physiol. 2019;10:1279. doi: 10.3389/fphys.2019.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin M., Zhang H., Wang J., Shao D., Yang H., Huang Q., et al. Response of intestinal metabolome to polysaccharides from mycelia of Ganoderma lucidum. Int J Biol Macromol. 2019;122:723–731. doi: 10.1016/j.ijbiomac.2018.10.224. [DOI] [PubMed] [Google Scholar]
- 35.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–U174. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burrello C., Garavaglia F., Cribiu F.M., Ercoli G., Lopez G., Troisi J., et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018:9. doi: 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin M., Kalainy S., Baskota N., Chiang D., Deehan E.C., McDougall C., et al. Faecal microbiota from patients with cirrhosis has a low capacity to ferment non-digestible carbohydrates into short-chain fatty acids. Liver Int. 2019;39(8):1437–1447. doi: 10.1111/liv.14106. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeSantis T.Z., Hugenholtz P., Keller K., Brodie E.L., Larsen N., Piceno Y.M., et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMurdie P.J. Holmes S: phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6) doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deehan E.C., Yang C., Perez-Munoz M.E., Nguyen N.K., Cheng C.C., Triador L., et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020;27(3):389-+ doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Martinez I., Lattimer J.M., Hubach K.L., Case J.A., Yang J.Y., Weber C.G., et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. Isme J. 2013;7(2):269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu J., Wang T.H., Xiao X., Cheng Y.Z., Wang F.Q., Jin M.L., et al. Clostridium butyricum ZJU-F1 benefits the intestinal barrier function and immune response associated with its modulation of gut microbiota in weaned piglets. Cells-Basel. 2021;10(3) doi: 10.3390/cells10030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aragon-Vela J., Solis-Urra P., Ruiz-Ojeda F.J., Alvarez-Mercado A.I., Olivares-Arancibia J. Plaza-diaz J: impact of exercise on gut microbiota in obesity. Nutrients. 2021;13(11) doi: 10.3390/nu13113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L., Lu X., Liu L., Voglmeir J., Zhong X., Yu Q. Akkermansia muciniphila protects intestinal mucosa from damage caused by S. pullorum by initiating proliferation of intestinal epithelium. Vet Res. 2020;51(1):34 doi: 10.1186/s13567-020-00755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lensu S., Pariyani R., Makinen E., Yang B., Saleem W., Munukka E., et al. Prebiotic xylo-oligosaccharides ameliorate high-fat-diet-induced hepatic steatosis in rats. Nutrients. 2020;12(11) doi: 10.3390/nu12113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An M., Park Y.H., Lim Y.H. Antiobesity and antidiabetic effects of the dairy bacterium Propionibacterium freudenreichii MJ2 in high-fat diet-induced obese mice by modulating lipid metabolism. Sci Rep. 2021;11(1):2481. doi: 10.1038/s41598-021-82282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen N.K., Deehan E.C., Zhang Z.X., Jin M.L., Baskota N., Perez-Munoz M.E., et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang C.J., Lin C.S., Lu C.C., Martel J., Ko Y.F., Ojcius D.M., et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. 2015:6. doi: 10.1038/ncomms8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao J.N., Shi W., Zhang J.Q., Zhang X.P., Zhang H., Wang Z.Y., et al. Response surface methodology for the fermentation of polysaccharides from Auricularia auricula using Trichoderma viride and their antioxidant activities. Int J Biol Macromol. 2020;155:393–402. doi: 10.1016/j.ijbiomac.2020.03.183. [DOI] [PubMed] [Google Scholar]
- 52.Kong X.H., Duan W.W., Li D.J., Tang X.X., Duan Z.H. Effects of polysaccharides from Auricularia auricula on the immuno-stimulatory activity and gut microbiota in immunosuppressed mice induced by cyclophosphamide. Front Immunol. 2020:11. doi: 10.3389/fimmu.2020.595700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen N.N., Zhang H., Zong X., Li S.Y., Wang J.J., Wang Y.Z., et al. Polysaccharides from Auricularia auricula: Preparation, structural features and biological activities. Carbohyd Polym. 2020:247. doi: 10.1016/j.carbpol.2020.116750. [DOI] [PubMed] [Google Scholar]
- 54.Isken F., Klaus S., Osterhoff M., Pfeiffer A.F.H., Weickert M.O. Effects of long-term soluble vs. insoluble dietary fiber intake on high-fat diet-induced obesity in C57BL/6J mice. J Nutr Biochem. 2010;21(4):278–284. doi: 10.1016/j.jnutbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Lam K.C., Araya R.E., Huang A., Chen Q.Y., Di Modica M., Rodrigues R.R., et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338-+ doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anhe F.F., Nachbar R.T., Varin T.V., Trottier J., Dudonne S., Le Barz M., et al. Treatment with camu camu (Myrciaria dubia) prevents obesity by altering the gut microbiota and increasing energy expenditure in diet-induced obese mice. Gut. 2019;68(3):453–464. doi: 10.1136/gutjnl-2017-315565. [DOI] [PubMed] [Google Scholar]
- 57.Defnoun S., Labat M., Ambrosio M., Garcia J.L., Patel B.K.C. Papillibacter cinnamivorans gen. nov., sp nov., a cinnamate-transforming bacterium from a shea cake digester. Int J Syst Evol Micr. 2000;50:1221–1228. doi: 10.1099/00207713-50-3-1221. [DOI] [PubMed] [Google Scholar]
- 58.Shen L. Gut, oral and nasal microbiota and Parkinson's disease. Microb Cell Fact. 2020;19(1) doi: 10.1186/s12934-020-01313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J., Zhang B.R., Wei G.F., Pang X., Wei H., Li M., et al. Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl Environ Microb. 2006;72(8):5232–5238. doi: 10.1128/AEM.00151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bendtsen L.Q., Blaedel T., Holm J.B., Lorenzen J.K., Mark A.B., Kiilerich P., et al. High intake of dairy during energy restriction does not affect energy balance or the intestinal microflora compared with low dairy intake in overweight individuals in a randomized controlled trial. Appl Physiol Nutr Me. 2018;43(1):1–10. doi: 10.1139/apnm-2017-0234. [DOI] [PubMed] [Google Scholar]
- 61.Roses C., Cuevas-Sierra A., Quintana S., Riezu-Boj J.I., Martinez J.A., Milagro F.I., et al. Gut microbiota bacterial species associated with mediterranean diet-related food groups in a northern Spanish population. Nutrients. 2021;13(2) doi: 10.3390/nu13020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verdam F.J., Fuentes S., de Jonge C., Zoetendal E.G., Erbil R., Greve J.W., et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity. 2013;21(12):E607–E615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 63.Hermes G.D.A., Reijnders D., Kootte R.S., Goossens G.H., Smidt H., Nieuwdorp M., et al. Individual and cohort-specific gut microbiota patterns associated with tissue-specific insulin sensitivity in overweight and obese males. Sci Rep-Uk. 2020;10(1) doi: 10.1038/s41598-020-64574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Labus J.S., Hollister E.B., Jacobs J., Kirbach K., Oezguen N., Gupta A., et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017:5. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uchimura K., Hayata M., Mizumoto T., Miyasato Y., Kakizoe Y., Morinaga J., et al. The serine protease prostasin regulates hepatic insulin sensitivity by modulating TLR4 signalling. Nat Commun. 2014:5. doi: 10.1038/ncomms4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qatanani M., Lazar M.A. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Gene Dev. 2007;21(12):1443–1455. doi: 10.1101/gad.1550907. [DOI] [PubMed] [Google Scholar]
- 67.Caricilli A.M., Picardi P.K., de Abreu L.L., Ueno M., Prada P.O., Ropelle E.R., et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. Plos Biol. 2011;9(12) doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Chan C.C., Harley I.T.W., Pfluger P.T., Trompette A., Stankiewicz T.E., Allen J.L., et al. A BAFF/APRIL axis regulates obesogenic diet-driven weight gain. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-23084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abumrad N.A. The liver as a hub in thermogenesis. Cell Metab. 2017;26(3):454–455. doi: 10.1016/j.cmet.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 70.Rial S.A., Jutras-Carignan A., Bergeron K.F. Mounier C: A high-fat diet enriched in medium chain triglycerides triggers hepatic thermogenesis and improves metabolic health in lean and obese mice. Bba-Mol Cell Biol L. 2020;1865(3) doi: 10.1016/j.bbalip.2019.158582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.