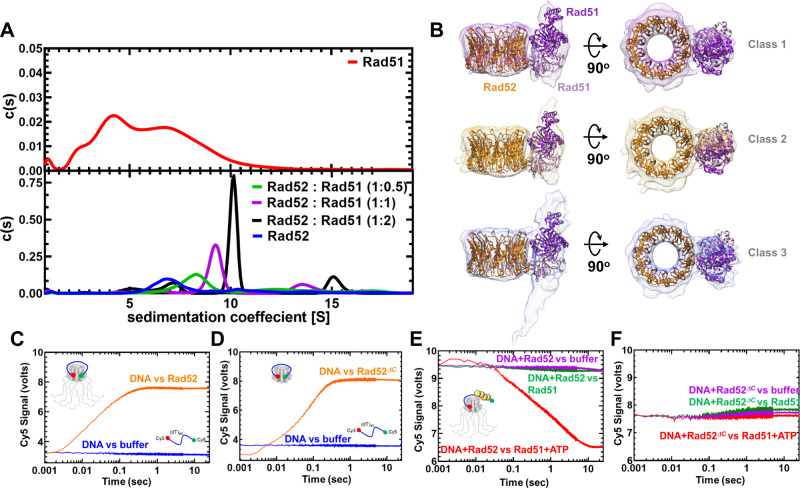
Fig. 7. Rad51 interacts with two distinct regions in Rad52.
A AUCSV analysis (top panel) shows polydispersity of Rad51 with multiple oligomeric states. When increasing amounts of Rad51 (1, 10, or 10 µM) are mixed with a fixed concentration of Rad52 (10 µM monomer), formation of Rad52-Rad51 complexes is observed. The size of the complexes increases as a function of Rad51 concentration in the solution. B 3D volumes from cryo-EM analysis of Rad52-Rad51 are shown. Structures of Rad52 and Rad51 (1SZP) are fit into the density. 3D volumes generated from three different classes are shown. The data fit well to a dimer of Rad51 bound (ordered) bound to one region in the Rad52 ring. Given the low resolution of the maps, Rad51 can be positioned in multiple orientations in the complex. Higher resolution structures will be required to better understand the details of the interaction. C Stopped flow FRET experiments were performed with a (dT)97 ssDNA labeled at the ends with Cy3 (3′; donor) and Cy5 (5′; acceptor). In the absence of protein, no changes in FRET are observed (blue) and a robust increase is captured when Rad52 is introduced (orange). Similar experiments done with D Rad52ΔC show wrapping of ssDNA. E FRET experiments were performed using prewrapped Rad52-ssDNA complexes and change in FRET was measured upon addition of Rad51 in the absence (green) or presence (red) of ATP. A loss in FRET is observed when Rad51 filaments are formed on the ssDNA in the presence of ATP. F Similar experiments performed with Rad52ΔC do not show a decrease in FRET suggesting an impediment to Rad51 filament formation. Source data are provided as a Source data file.