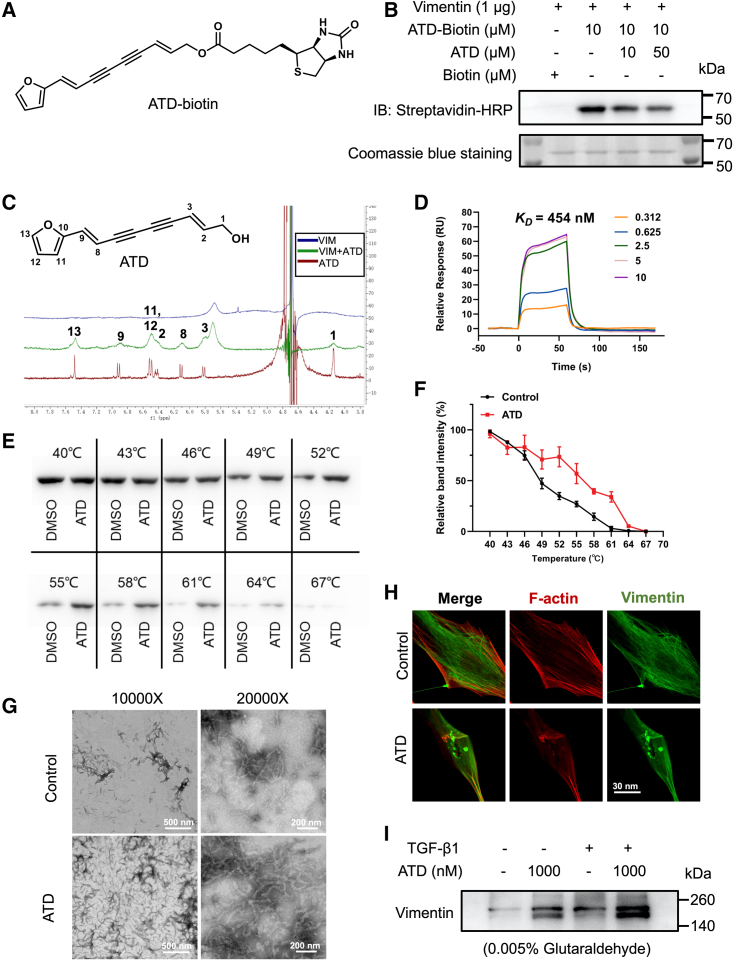
Figure 5.
ATD targets the intermediate filament protein VIM
(A) ATD-biotin chemical structures. (B) Purified His-VIM was incubated with the indicated concentrations of ATD for 1 h followed by incubation with ATD-biotin (10 μM) for 1 h at 37°C. ATD-biotin bound to purified VIM was detected after SDS-PAGE with streptavidin-HRP. (C) Ligand-observed 1D 1H-NMR of the peaks of 2 mM ATD (red) becomes wider (green) in the presence of 20 μM VIM. Positive and negative signals identify VIM binding and non-interacting molecules, respectively. (D) Surface plasmon resonance sensorgrams for the binding of ATD to VIM. (E) The detectable remaining amount of VIM after ATD treatment varies with the temperature used for heat treatment of HFL1 cells. (F) CETSA melt curve quantified by western blotting (n = 3). (G) VIM was polymerized in the presence of 5 mM MgCl2, 100 mM KCl, and 170 mM NaCl by incubation at 37°C for 1 h. The protein was fixed with 0.5% glutaraldehyde, stained with uranyl acetate, and observed by transmission electron microscopy. Polymerization of tetrameric VIM in the presence of 20 μM ATD produces extensive filamentous aggregates. (H) HFL1 cells treated with DMSO or 1 μM ATD for 24 h were stained for VIM with anti-VIM antibody (green) and co-stained with F-actin (red). The presence of numerous cytoplasmic particulate granules that co-stain for VIM and disrupted F-actin in ATD-treated cells compared with control. Scale bar, 30 μm. (I) Cell lysates treated with or without ATD samples were crosslinked with 0.005% glutaraldehyde, and the VIM forms were detected by western blotting (n = 3). The data are presented as the mean ± SEM.