Abstract
Purpose
We aimed to evaluate the time and cost of developing prompts using large language model (LLM), tailored to extract clinical factors in breast cancer patients and their accuracy.
Materials and Methods
We collected data from reports of surgical pathology and ultrasound from breast cancer patients who underwent radiotherapy from 2020 to 2022. We extracted the information using the Generative Pre-trained Transformer (GPT) for Sheets and Docs extension plugin and termed this the “LLM” method. The time and cost of developing the prompts with LLM methods were assessed and compared with those spent on collecting information with “full manual” and “LLM-assisted manual” methods. To assess accuracy, 340 patients were randomly selected, and the extracted information by LLM method were compared with those collected by “full manual” method.
Results
Data from 2,931 patients were collected. We developed 12 prompts for Extract function and 12 for Format function to extract and standardize the information. The overall accuracy was 87.7%. For lymphovascular invasion, it was 98.2%. Developing and processing the prompts took 3.5 hours and 15 minutes, respectively. Utilizing the ChatGPT application programming interface cost US $65.8 and when factoring in the estimated wage, the total cost was US $95.4. In an estimated comparison, “LLM-assisted manual” and “LLM” methods were time- and cost-efficient compared to the “full manual” method.
Conclusion
Developing and facilitating prompts for LLM to derive clinical factors was efficient to extract crucial information from huge medical records. This study demonstrated the potential of the application of natural language processing using LLM model in breast cancer patients. Prompts from the current study can be re-used for other research to collect clinical information.
Keywords: Automatic data processing, Artificial intelligence, Natural language processing, Breast cancer, Clinical reports
Introduction
In radiation therapy for breast cancer patients, numerous clinical factors are considered. For instance, when the National Comprehensive Cancer Network panels [1] recommend comprehensive regional nodal irradiation for pN1 patients, it also recommends considering clinical factors such as whether the primary tumor is small or there is only one metastasis. Indeed, clinical decision to irradiate full regional lymph nodes or not depend on clinical factors including age, nuclear grade, molecular subtype, resection margin status, lymphovascular invasion and extranodal extension [2–5] as well as TNM stage. Thus, pathologic factors and radiologic findings are carefully reviewed to make an optimal decision in clinical practice. Radiation oncologists manually reviewed the medical record of each patient, seeking important factors to support their decision. When collecting clinical information for research work, medical reports are thoroughly reviewed, factors are manually classified, and the case report form is constructed. This process is labor-intensive, costly, and time-consuming. However, automation of these processes has been challenging due to difficulties in processing unstructured, narrative-style reports generated from diagnostic work-up for breast cancer patients.
Recent advances in natural language processing (NLP) play a pivotal role in solving complex problems like the challenges mentioned above. NLP makes it possible to reliably extract important information from free text, and many fields of medicine, including radiation oncology, could benefit from these techniques [6]. In the field of NLP, state-of-the-art large language models (LLMs) like GPT-4 (Generative Pre-trained Transformer 4) have shown outstanding abilities in understanding and generating human language [7]. This enables LLMs to comprehend textual data and establish contextual connections, leading to revolutionary achievements. Further, this capability allows LLMs to analyze complex medical data, extract crucial information, and support decision-making. As such, the effective utilization of LLMs offers tremendous potential in the automation of traditional medical chart review.
To use LLM effectively, “prompts” must be properly developed. Prompts are input sentences or phrases for LLM to perform a specific action. The content and structure of these prompts greatly influence the output and performance of LLMs [8]. Depending on the prompt, LLM model determine which information to process and what type of answers to generate. Thus, design of proper prompts is key to maximizing the capabilities of LLMs, given that inappropriate prompts can cause models to misunderstand or produce unexpected results. In particular, in the field of radiation oncology, research on prompt engineering is required for LLM to extract accurate information from unstructured medical reports.
In this study, prompts were developed to extract the required information using LLM from surgical pathology reports and preoperative ultrasound reports of breast cancer patients. The time and cost needed to develop the prompt was assessed and compared with manual methods. Also, the accuracy of the extracted information was evaluated by comparing it with information collected manually.
Materials and Methods
We collected data from breast cancer patients who received postoperative radiotherapy (RT) from 2020 to 2022 in our institution. Male breast cancer patients, patients who received palliative RT, and patients who did not undergo surgery at our institution were excluded. For the study population, the findings from the earliest breast ultrasound within one year before the surgery and the report from surgical pathology were collected. The overall study schema is depicted in Fig. 1.
Fig. 1.
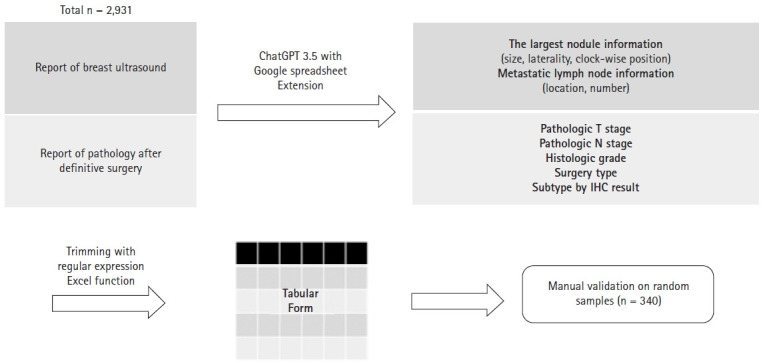
The schema of the current study. Using ChatGPT 3.5 model, information about clinical T and N stage was extracted from ultrasound readings, and pathologic T and N stage and additional factors were extracted from pathology readings. Then, trimming was performed and organized in tabular form. For validation, a sample was randomly selected to evaluate the accuracy. GPT, Generative Pre-trained Transformer; IHC, immunohistochemistry.
As the target LLM, we selected the ChatGPT and accessed it with an extension program of Google Sheets, named as GPT for Sheets and Docs (https://gptforwork.com). This plugin, developed by Talarian, is an application that allows the GPT model to be used directly in Google Sheets and provides additional custom features. This makes using various LLM models such as GPT3 and ChatGPT models in Google Sheets possible. In this study, we used the ChatGPT (gpt-3.5-turbo) model.
From the ultrasound reports, prompts were designed to extract and organize factors related to the clinical stage. To determine the clinical T stage, we designed prompts to extract the size and location of the largest suspected cancerous mass within the breast. To extract information about the clinical N stage, we designed prompts to extract the number of suspected metastatic lymph nodes for each nodal area. In addition, information about laterality and tumor location was also extracted.
In the surgical pathology report, prompts were designed to extract factors like tumor size and number of metastatic lymph nodes, which determines the pathological stage. We also designed prompts to extract clinical factors such as histologic grade, neoadjuvant chemotherapy status, resection margin status, molecular subtype, lymphovascular invasion, and extracapsular extension. In addition, we designed prompts to extract factors, such as surgery type and metastatic lymph node ratio.
To evaluate the efficiency and accuracy of the extraction method using LLM, we compared three methods of collecting clinical information from medical records. First, the “full manual” method was done by JYS, a resident physician in Radiation Oncology. The method is a way to manually collect information from the medical records of each patient, and to ensure 100% accuracy, the information was verified by another physician after collection. Second, the “LLM-assisted manual” method was done by the same resident physician who did the “full manual” method, but with the assistance of the information already collected using LLM. Lastly, the “LLM” method used LLM alone to extract information of clinical factors. To establish comparability throughout the three methods, 340 patients, were randomly selected using the RAND function in Microsoft Excel to extract information. We calculated the sample size to represent the accuracy of the entire 2,913 cases with a confidence level of 95% and a margin of error of 5%.
The accuracy and time- and cost-efficiency were assessed using the following methods. First, the accuracy of the “LLM” method was assessed by regarding the information collected by the “full manual” method as the ground truth and comparing the data collected with both methods. The accuracy rate was calculated by each factor. Also, the time spent to collect the information, to design prompts, or to trim information collected by LLM was measured. Since only 340 patients were included in “full manual” and ‘LLM-assisted manual’ methods, a factor of 8.57 (2,931/340) was multiplied by the measured time to extrapolate, and establish comparability between the three methods. Furthermore, the cost spent to collect the information was estimated. In the “full manual” and “LLM-assisted manual” methods, the cost was estimated by multiplying the measured time by US $7.4 per hour, which is South Korea’s minimum wage in 2023. In the “LLM-assisted manual” and “LLM” methods, the estimated cost for prompt design and the GPT application programming interface (API) fee was measured. Since the “LLM” method was performed on entire patients, the cost for 340 patients was calculated by dividing the total GPT API fee by 8.57.
Results
In total, 2,931 breast cancer patients treated at our institution were included in this study. Table 1 shows the factors, function types, prompts designed, and their corresponding results in this study. A representative example of a surgical pathology report and an ultrasound report could be found in Supplementary Figs. S1 and S2.
Table 1.
The factors, function types, prompts designed, and their corresponding results
| Factors | Function type | Prompt | Result | |
|---|---|---|---|---|
| Ultrasound reports | Largest nodule information | Extract | Information about largest nodule in diameter or mass with BI-RADS classification of C4 or higher. if there is no C4 or higher nodule, just say 'no cancer'. if not 'no cancer', answer form is 'longest diameter (e.g., 1.0 cm, 2.5 cm by cm), laterality (e.g., Rt/Lt), orientation (by clockface, e.g., 1H, 11.5H) or by quadrant (e.g., SA, center, UO, IL), BI-RADS classification'. all answer is in a single line. | 2.3, Lt, 2H, C5/6 |
| Size | Format | Longest one direction diameter. say just number without unit, not other information. If the unit is mm, change it to cm. | 2.3 | |
| Laterality | Format | Laterality by Lt or Rt. only answer Lt or Rt | Lt | |
| Clockwise orientation | Format | Orientation by clockface, like 1H, 2H, 10.5H | 2 | |
| Metastatic lymph node | Extract | This is sono reading. from now on, you are radiologist, count number of suspicion or enlarged lymph nodes. do not count suspicion breast nodule. do things step by step. | Inner : N/A / axillary : 5 / IMN : N/A / SCL : N/A | |
| Answer form 'inner: ## / axillary : ## / IMN : ## / SCL : ##' answer in single line. 'inner' means intramammary lymph nodes. 'IMN' means internal mammary lymph nodes. 'SCL' mean supra clavicular lymph nodes. | ||||
| Pathology reports | Pathological T stage | Extract | The invasive tumor size 또는 종괴의 크기 as a long diameter 또는 the extent of in situ. | 1.5 × 0.4 × 2.0 cm (invasive tumor size) |
| Format | T stage. You are breast cancer pathologist interpreting reports with AJCC 8th staging system | T1c | ||
| Histologic grade | Extract | The histologic grade written in pathologic reports | II/III | |
| Format | Answer just number according to histologic grade. You are breast cancer pathologist | 2 | ||
| Surgery type | Extract | Surgery type | Breast conserving surgery | |
| Pathological N stage | Extract | The number of positive or metastatic lymph nodes out of total dissected lymph nodes. If nodes were not dissected or not submitted, just say 'not submitted' | Number of metastatic lymph nodes: 1 out of 11 examined lymph nodes. | |
| Format | N stage. You are breast cancer pathologist interpreting reports with AJCC 8th staging system. If not submitted, just say 'Nx' | N1 | ||
| Metastatic lymph node ratio | Format | Just say only the number after calculating the ratio of the number of positive or metastatic lymph nodes by the total number of examined or dissected lymph nodes | 0.09 | |
| Lymph node sampling type | Format | Say SLNB if all examined or dissected lymph nodes were sentinel nodes. If not, say ALND | ALND | |
| Number of metastatic lymph nodes | Extract | Just say the number of metastatic or involved lymph nodes, not total harvested or dissected lymph nodes | 1 | |
| Neoadjuvant chemotherapy | Extract | Just say 'Yes' if this pathologic report described post neo-adjuvant chemotherapy or 'yp'stages according to AJCC staging system. If not, just say 'No'. | Yes (post-neoadjuvant chemotherapy status) | |
| Resection margin | Extract | Evaluate the margin status, reviewing this pathologic report. Answer in one of 3 words: 'Clear', 'Close', or 'Positive'. You are breast cancer pathologist. | Clear | |
| IHC result | Extract | Concatenate the immunohistochemistry results including estrogen, progesterone, Ki-67, and gene amplification results in single line. If there isn't, just say 'N/A'. Don't make multiple lines in answer. | Estrogen Receptor alpha, positive in 90%; progesterone receptor, positive in 40% (S 21-0030586) | |
| Estrogen Receptor alpha, positive in 90%; progesterone receptor, positive in 80% (S 21-0030587) | ||||
| Ki-67, positive in 1% | ||||
| Format | Just say 'Positive' if the expression of estrogen receptor is positive or more than 1+, based on AJCC breast cancer staging. Otherwise, just say 'No'. | Positive | ||
| Format | Just say 'Positive' if the expression of HER2 is more than 2+ or FISH amplification is positive, based on AJCC breast cancer staging. Otherwise, just say 'Negative'. | Negative | ||
| Format | Just extract the number how much percentage of Ki-67 expression is in positive. If Ki-67 was not found, just say 'N/A'. | 0.01 | ||
| Format | As shown in immunochemistry result, breast cancer is triple-negative type, then say 'Yes'. If not, just say 'No' | No | ||
| Lymphovascular invasion | Extract | The lymphatic invasion in pathologic report | Lymphatic emboli: present, minimal | |
| Extracapsular extension | Extract | The presence or absence or N/A of extracapsular extension in the pathology results | Extracapsular extension: N/A |
AJCC, American Joint Committee on Cancer; ALND, axillary lymph node dissection; BI-RADS, Breast Imaging Reporting and Data System; FISH, fluorescence in situ hybridization; HER2, human epidermal growth factor receptor 2; IL, inferio-lateral; IMN, internal mammary lymph node; N/A, not accessible; SA, subareolar; SCL, supraclavicular lymph node; SLNB, sentinel lymph node biopsy; UO, upper-outer.
Prompts were developed using the Extract function and the Format function from GPT for Sheets and Docs. The Extract function extracts the required information from the reports. The Format function was used to convert them into a structured format. For future statistical analysis, it is essential to standardize the format of responses. Therefore, the choice or type of response was predetermined through the prompts. When the required information could not be extracted at once, the extraction was performed in two steps. For example, when the information regarding the largest nodule among the nodes with suspected malignancy is needed, the information of the node that corresponds to the condition is first extracted, and then the information on diameter, laterality, and clockwise location is extracted again from that extracted information.
In some cases, simple computations to trim the extracted information were necessary. These were performed using IF, AND, and OR functions in Microsoft Excel. For instance, functions were used to determine the breast cancer subtype based on immunohistochemistry results or to identify whether the breast cancer was located on the inner or outer side based on the laterality and clockwise location. When additional trimming was needed for statistical analysis, the information was trimmed using regular expressions in Python.
The accuracy of the information extraction by LLM was calculated by each factor and is shown in Table 2. Regarding all the factors, the average accuracy was 87.7%, which could be translated to roughly 298 out of 340 patients. Among all factors, lymphovascular invasion had the highest accuracy of 98.2%. In contrast, neoadjuvant chemotherapy status and tumor location had the lowest accuracy of 47.6% (162 out of 340) and 63.8% (217 out of 340), respectively.
Table 2.
The accuracy of the data extraction by LLM
| Factor | Correct | Total | Accuracy (%) |
|---|---|---|---|
| Clinical T stage | 279 | 340 | 81.9 |
| Clinical N stage | 311 | 340 | 91.5 |
| Tumor location | 217 | 340 | 63.8 |
| Surgery type | 318 | 340 | 93.4 |
| Neoadjuvant chemotherapy | 162 | 340 | 47.6 |
| Pathologic T stage | 295 | 340 | 86.7 |
| Histologic grade | 334 | 340 | 98.1 |
| Lymphovascular invasion | 334 | 340 | 98.2 |
| Resection margin | 298 | 340 | 87.7 |
| Lymph node sampling type | 295 | 340 | 86.7 |
| Pathologic N stage | 314 | 340 | 92.4 |
| Metastatic lymph node ratio | 324 | 340 | 95.3 |
| Extracapsular extension | 305 | 340 | 89.6 |
| Estrogen receptor | 324 | 340 | 95.3 |
| HER2 | 327 | 340 | 96.3 |
| Ki-67 | 311 | 340 | 91.5 |
| Triple-negative breast cancer | 324 | 340 | 95.3 |
| Overall (average) | 298 | 340 | 87.7 |
LLM, large language model; HER2, human epidermal growth factor 2.
The time- and cost-efficiency of the information extraction by LLM were also assessed. Regarding the time-efficiency, it took 1.5 hours for designing the prompts for the ultrasound report and 2 hours for the surgical pathology report. Responses to the prompts were outputted in parallel for each cell in the table via the GPT server. The entire response output process took 15 minutes. Trimming the data took approximately 30 minutes in total, which was mostly coding time, and the actual application was completed in a few seconds. In total, approximately 4 hours were needed in extracting clinical factors from 2,931 breast cancer patients. In terms of cost, using the GPT model through the GPT API incurs a fee per token. In the current study, US $6.04 for ultrasound interpretation and US $59.76 for surgical pathology interpretation was charged using the GPT API, for a total of US $65.8. Also, the whole process took 4 hours to design the prompts and trim the data, which could be translated to a wage cost of US $29.6, when applying the minimum wage of South Korea in 2023. The response output process was excluded from the wage cost calculation because the process could be done automatically.
The time and cost spent on collecting the information using “full manual,” “LLM-assisted manual,” and “LLM” methods were measured and estimated for comparison (Table 3). For all 2,913 patients, the time spent for the “full manual,” “LLM-assisted manual,” and “LLM” methods were 122.6 hours, 79.4 hours, and 4 hours, respectively. The estimated cost for “full manual,” “LLM-assisted manual,” and “LLM” methods were US $909.3, $653.4, and $95.4, respectively. By using “LLM-assisted manual” and “LLM” methods compared to the “full manual” method, we could save 43.2 hours and US $255.9, and 118.6 hours and US $813.9 in all 2,913 patients, respectively.
Table 3.
The time and cost spent on collecting the data using “full manual,” “LLM-assisted manual,” and “LLM” methods
| Full manual | LLM-assisted manual | LLM | ||||
|---|---|---|---|---|---|---|
| 340 patients | All patients (extrapolated) | 340 patients | All patients (extrapolated) | 340 patients | All patients | |
| Time spent (hr) | ||||||
| For data collection | 14.3 | 122.6 | 8.8 | 75.4 | - | - |
| For prompt design and trimming | - | - | 4.0 | 4.0 | 4.0 | 4.0 |
| Total | 14.3 | 122.6 | 12.8 | 79.4 | 4.0 | 4.0 |
| Time saved compared to “full manual” | - | - | 1.5 | 43.2 | 9.3 | 118.6 |
| Estimated cost (US dollar) | ||||||
| Manual data collection wage cost | 106.1 | 909.3 | 65.6 | 558 | - | - |
| Prompt design and trimming wage cost | - | - | 29.6 | 29.6 | 29.6 | 29.6 |
| GPT API usage fee | - | - | 7.7 | 65.8 | 7.7 | 65.8 |
| Total | 106.1 | 909.3 | 102.9 | 653.4 | 37.3 | 95.4 |
| Cost saved compared to “full manual” | - | - | 3.2 | 255.9 | 68.8 | 813.9 |
API, application programming interface; LLM, large language model; Bold, total time and cost.
Discussion and Conclusion
In the current study, we investigated the efficiency and accuracy of the utilization of LLM in extracting RT-related factors. From 2,931 breast cancer patients, we extracted clinical factors from the reports of ultrasound and surgical pathology. The whole process took 4 hours and cost US $95.4, and the average accuracy was 87.7%.
The GPT model, which is employed in the current study, is intended to imitate natural conversations, and does not contain logical thinking [9]. Thus, it does not understand the meaning of sentences but is merely aligning the most likely word to use. In other words, there is no logic in the response it gives. For example, in the current study, we first wanted to separate the location of a breast tumor into inner and outer based on laterality and the clockwise direction from the nipple. Despite many trials and errors, we failed to implement the function in a GPT model. This may be because the GPT model failed to comprehend the logic that the clockwise direction is opposite according to the laterality of the breast in separating inner from outer lesions. Also, another well-known feature of the GPT model is a phenomenon called the hallucination [10]. This is a phenomenon that the model responds with a plausible answer that is incorrect. For example, when the GPT-3.5 is asked “Explain to me the clinical N stage of breast cancer according to the American Joint Committee on Cancer (AJCC) 8th edition,” it comes up with a plausible answer which is an explanation about the pathological N stage. The hallucination was seen time to time in extracting information in the current study. For example, when asked about the diameter of the tumor from the ultrasound report, it sometimes responded with the distance from the nipple. We speculate that this hallucination occurred because the two values are both written in numerical values in centimeter. In another example, the category “Neoadjuvant chemotherapy” was accurate at less than 50%. It could be due to hallucination. Even though there was no information about neoadjuvant chemotherapy in the pathology report, LLM gave a yes or no answer instead of saying that there was insufficient information. A deep understanding in such features of LLMs is crucial to designing prompts and applying it to use.
Developing an effective prompt to get the desired outcome, which is called prompt engineering, is the most crucial point in utilizing LLM. In prompt engineering, the features of LLMs should be well understood by developers. When using an LLM, users may avoid using jargon, instead, should provide all the information needed to generate a response. Also, it is recommended to write the logic leading up to the desired outcome in the prompt, rather than trying to achieve the desired outcome all at once, thereby, the LLM can follow the logical process. For example, in interpreting an ultrasonography report, instead of saying, “Identify the clinical T stage,” it is better to say, “Here's an ultrasound reading of a breast cancer patient, find the mass with the largest diameter and tell me its diameter in centimeters.” Also, these results can be improved with a well-known few-shot learning or fine-tuning method by introducing an example within the prompt and having the LLM replicate the logical flow [11]. Since we could not fully predict the output of LLM, it is essential to modify and correct the prompt through a trial-and-error method, rather than completing it at once. Understanding these features will provide appropriate answers to users when effective prompt engineering is adopted in coding clinical data.
Information extraction by developing prompts from LLM is extremely efficient in terms of both time and expenses, especially when it is applied to a task handling huge data. From raw data of thousand patients to well-organized spreadsheet data, we could save 118.6 hours and US $813.9 by using “LLM” method. After completing the prompt design, the cost of developing the prompt is fixed, which could save expenses for research that needs to be encoded from large data, such as pathological or radiographic findings of patients with breast or prostate cancer treated with radiation therapy. This method is a much-awaited in labor market like South Korea, where the hiring of qualified healthcare provider is expensive and scarce. For the last decade in South Korea, there has been a steep growth in wages with the minimum wage almost doubling [12], and that of the healthcare providers are no exception [13]. In addition, the “Act on the Improvement of Training Conditions and Status of Medical Residents” was enacted in 2015, restricting the working hours of medical residents [14]. Due to advantage of the using LLM in large-scale tasks, development, and facilitating prompts are employed in other expert fields as well [15].
The accuracy of the information extracted by prompts using LLM was 87.7%, which is an encouraging result compared to conventional NLP models. Juhn et al. [16] reported 80%–98% accuracy in identifying the presence or severity of allergic conditions through medical record review using an NLP-based model [16]. In addition, Tang et al. [17] reported a low accuracy of 23.4% in biomedical named entity recognition using the ChatGPT, and a higher accuracy of 75.9% in LLM for medical tasks. Although the accuracy of 87.7% in the current study is notable, it should be cautioned to use without supervision. Manual examination should be done to re-examine the extracted information. Alternatively, a hybrid method that we named the “LLM-assisted manual” method could be a reasonable way to compromise in a real-world setting. By referring to the information extracted, one can collect information more efficiently, while still maintaining the level of accuracy of a manual information collection. Recently, the GPT-4 was released and expected to reduce the frequency of hallucinations and improve accuracy [7]. We expect that LLMs specialized in medical tasks may elevate the accuracy of extraction even better [18,19].
There were several limitations in this study. First, the clinical N stage did not strictly follow the AJCC staging system. The information in the ultrasound report was insufficient to extract whether a lymph node is movable or not. Thus, distinguishing between clinical N1 and N2 solely depended on the number of nodal metastasis or the presence of an internal mammary lymph node metastasis without axillary metastasis, and has discrepancy with the AJCC staging system. Also, the “full manual” method, which is used as a ground truth, is not perfect. Although manual process is the control for the “LLM” process, the “full manual” method does not guarantee 100% accuracy in information collection due to the human error. It could be improved if two or more people could cross-check each other’s collected information to approach 100% accuracy for a solid ground truth. Moreover, using minimum wage in the evaluation of total cost may be inaccurate. Healthcare providers such as doctors or nurses, who would likely collect the clinical information from the medical records in real-life, receive more than a minimum wage. Therefore, the exact cost of manual information collection could be higher. Finally, the extrapolation of time and cost taken in 60 patients into 2,913 patients may be inaccurate. Validation with more data is required.
In conclusion, we showed that using LLM prompts is an efficient way to extract crucial information from the medical records of breast cancer patients and to construct well-fined clinical data. This method is expected to save lots of effort from daily practice and research work. Prompts from the current study can be re-used for other investigators to collect clinical information.
Footnotes
Statement of Ethics
This study was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. H-2304-072-1422).
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Funding
This work was supported by the National R&D Program for Cancer Control through the National Cancer Center (NCC) funded by the Ministry of Health & Welfare, Republic of Korea (No. HA22C0044) to Kyung Hwan Shin and the Ministry of Science and Information & Communication Technology (No. NRF-2022R1F1A106345712) to Bum-Sup Jang.
Author Contributions
Conceptualization, Choi HS, Song JY, Jang BS. Funding acquisition, Shin KH, Jang BS. Investigation and methodology, Choi HS, Song JY, Jang BS. Project administration, Choi HS. Resources, Shin KH, Chang JH, Jang BS. Supervision, Jang BS. Writing of the original draft, Choi HS, Song JY. Writing of the review and editing, Choi HS, Song JY. Software, Choi HS. Validation, Choi HS. Formal analysis, Choi HS, Song JY. Data curation, Choi HS. Visualization, Choi HS.
Data Availability Statement
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but are available from Bum Sup Jang upon reasonable request.
Supplementary Materials
Supplementary materials can be found via https://doi.org/10.3857/roj.2023.00633.
A representative example of an ultrasound report.
A representative example of a surgical pathology report.
References
- 1.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology: breast cancer [Internet]. Plymouth Meeting, PA: National Comprehensive Cancer Network; 2023 [cited 2023 Sep 13]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf.
- 2.Park HJ, Shin KH, Kim JH, et al. Incorporating risk factors to identify the indication of post-mastectomy radiotherapy in N1 breast cancer treated with optimal systemic therapy: a multicenter analysis in Korea (KROG 14-23) Cancer Res Treat. 2017;49:739–47. doi: 10.4143/crt.2016.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada A, Hayashi N, Kumamaru H, et al. Prognostic impact of postoperative radiotherapy in patients with breast cancer and with pT1-2 and 1-3 lymph node metastases: a retrospective cohort study based on the Japanese Breast Cancer Registry. Eur J Cancer. 2022;172:31–40. doi: 10.1016/j.ejca.2022.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Jwa E, Shin KH, Lim HW, et al. Identification of risk factors for locoregional recurrence in breast cancer patients with nodal stage N0 and N1: who could benefit from post-mastectomy radiotherapy? PLoS One. 2015;10:e0145463. doi: 10.1371/journal.pone.0145463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viani GA, Godoi da Silva LB, Viana BS. Patients with N1 breast cancer: who could benefit from supraclavicular fossa radiotherapy? Breast. 2014;23:749–53. doi: 10.1016/j.breast.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Bitterman DS, Miller TA, Mak RH, Savova GK. Clinical natural language processing for radiation oncology: a review and practical primer. Int J Radiat Oncol Biol Phys. 2021;110:641–55. doi: 10.1016/j.ijrobp.2021.01.044. [DOI] [PubMed] [Google Scholar]
- 7.OpenAI GPT-4 technical report [Internet]. Ithaca, NY: arXiv.org; 2023 [cited 2023 Sep 13]. Available from: [DOI]
- 8.Clavie B, Ciceu A, Naylor F, Soulie G, Brightwell T. In: Natural language to information systems. Metais E, Meziane F, Sugumaran V, Manning W, Reiff-Marganiec S, editors. Cham, Switzerland: Springer; 2023. Large language models in the workplace: a case study on prompt engineering for job type classification; pp. 3–17. [Google Scholar]
- 9.Floridi L, Chiriatti M. GPT-3: its nature, scope, limits, and consequences. Minds Mach. 2020;30:681–94. [Google Scholar]
- 10.Lee P, Budeck S, Petro J, et al. Benefits, limits, and risks of GPT-4 as an AI chatbot for medicine. N Engl J Med. 2023;388:1233–9. doi: 10.1056/NEJMsr2214184. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Shi E, Yu S, et al. Prompt engineering for healthcare: methodologies and applications [Internet]. Ithaca, NY: arXiv.org; 2023 [cited 2023 Sep 13]. Available from: [DOI]
- 12.Seok BH, You HM. Macroeconomic impacts of increasing the minimum wage: the case of Korea. Econ Model. 2022;113:105880. [Google Scholar]
- 13.Minimum Wage Commission, Republic of Korea Announcement of the Results of the Healthcare Workforce Status Survey [Internet]. Sejong, Korea: Minimum Wage Commission; c2022 [cited 2023 Sep 13]. Available from: https://www.minimumwage.go.kr/minWage/policy/decisionMain.do.
- 14.Sohn S, Seo Y, Jeong Y, Lee S, Lee J, Lee KJ. Changes in the working conditions and learning environment of medical residents after the enactment of the Medical Resident Act in Korea in 2015: a national 4-year longitudinal study. J Educ Eval Health Prof. 2021;18:7. doi: 10.3352/jeehp.2021.18.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Heerden AC, Pozuelo JR, Kohrt BA. Global mental health services and the impact of artificial intelligence-powered large language models. JAMA Psychiatry. 2023;80:662–4. doi: 10.1001/jamapsychiatry.2023.1253. [DOI] [PubMed] [Google Scholar]
- 16.Juhn Y, Liu H. Artificial intelligence approaches using natural language processing to advance EHR-based clinical research. J Allergy Clin Immunol. 2020;145:463–9. doi: 10.1016/j.jaci.2019.12.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang R, Han X, Jiang X, Hu X. Does synthetic data generation of LLMs help clinical text mining? [Internet]. Ithaca, NY: arXiv.org; 2023 [cited 2023 Sep 13]. Available from: [DOI]
- 18.Kung TH, Cheatham M, Medenilla A, et al. Performance of ChatGPT on USMLE: potential for AI-assisted medical education using large language models. PLOS Digit Health. 2023;2:e0000198. doi: 10.1371/journal.pdig.0000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singhal K, Tu T, Gottweis J, et al. Towards expert-level medical question answering with large language models [Internet]. Ithaca, NY: arXiv.org; 2023 [cited 2023 Sep 13]. Available from: [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A representative example of an ultrasound report.
A representative example of a surgical pathology report.


