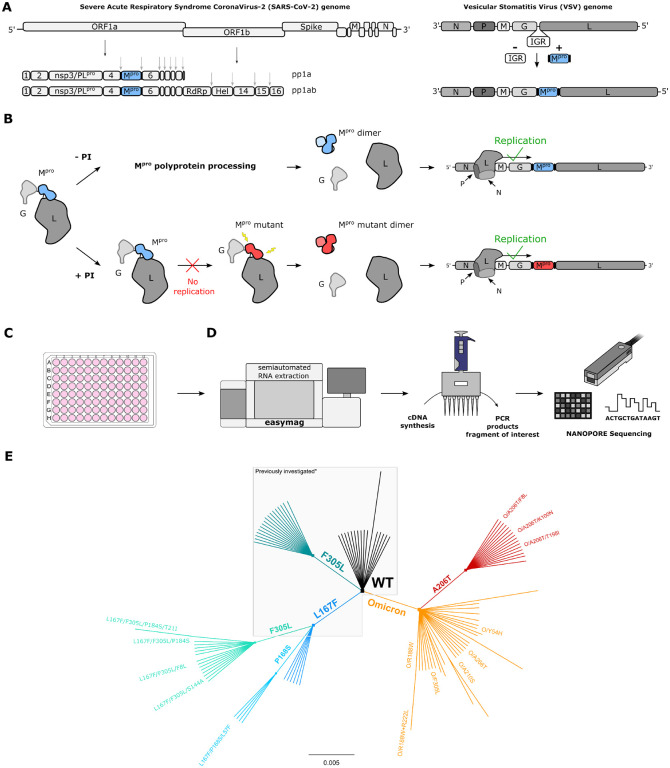
Fig. 1. VSV-G-Mpro-L construct: molecular mechanism, sequencing workflow, and mutant lineage phylogeny.
(A) Schematic representation of SARS-CoV-2 genome, polyproteins pp1a and pp1ab and the VSV-based mutation selection tool. The InterGenic Region (IGR) between proteins G and L of WT VSV was replaced with the SARS-CoV-2 Mpro Wuhan-1 sequence and its cognate autocleavage sites. Mpro genome positions in SARS-CoV-2 and in VSV-Mpro are highlighted in light blue. (B) The virus is fully dependent on Mpro for replication. Upon translation of G-Mpro-L, two outcomes are possible: 1. without an inhibitor, Mpro is free to process the polyprotein, and the transcription and replication complexes can assemble; 2. with an inhibitor, Mpro is inhibited, the polyprotein is not processed, and the virus is thus not able to replicate, unless it acquires a mutation rendering the Mpro less susceptible to the inhibitor. Then, the virus can replicate despite the inhibitor. (C) BHK21 cells in a 96-well plate are infected with VSV-Mpro. Here, the two outcomes described in panel b can occur. (D) Nanopore sequencing workflow. (E) Unrooted phylogenetic tree showing the relationship between original/parental viruses and mutants. Mpro variants belonging to the same parental virus are coloured accordingly: WT (black), F305L (sea green), L167F (blue), L167F/F305L (light sea green), L167F/P168S (light blue), Omicron (orange), Omicron/A206T (red). For clarity, only the names of the mutants investigated in this work are displayed. *Previously generated/investigated set of mutants in our first study (34).