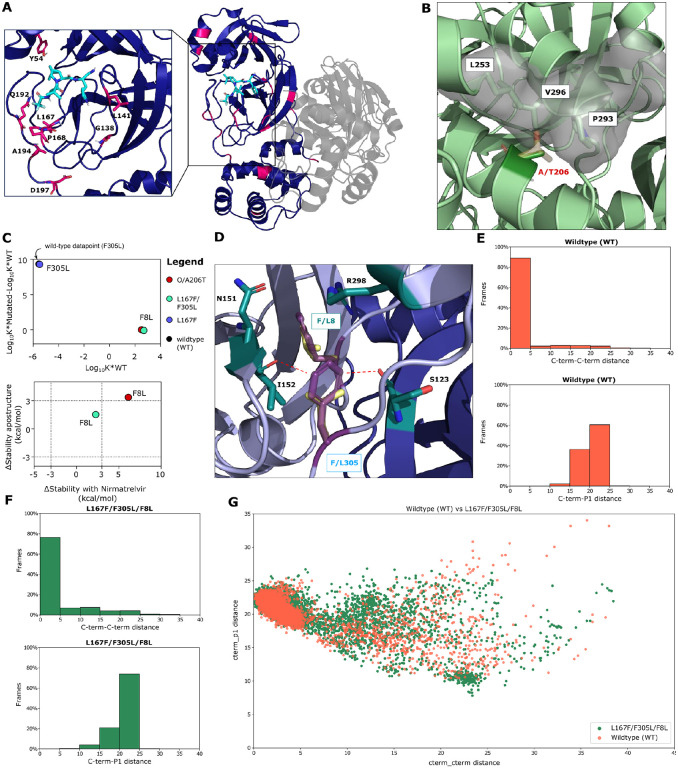
Fig. 5. Structural analysis of mutants.
(A) Overview of investigated mutants (pink) mapped onto the Mpro homodimer (dark violet and grey) bound to nirmatrelvir (light blue sticks, PDB 8DZ2) (48). Side chains of residues within the drug binding site are shown as sticks to highlight their distance to nirmatrelvir. (B) The WT alanine side chain of residue 206 (dark green sticks) is tightly packed against the hydrophobic side chains of residues L253, P293, and V296 (green sticks, PDB entry 7ALI) (51). The threonine side chain (yellow sticks) is polar and would clash with these residues as indicated by the light grey surface. (C) Dimerization affinity plot of F8L and F305L mutants (top) and Δ_Stability values (bottom) of apo and nirmatrelvir bound structures for the F8L mutants within different variant backgrounds. The additional dotted lines at x = ± 3 and y = ± 3 in the stability plot are cut-off values that we considered as a meaningful difference in protease stability (a positive value indicates a decrease in protease stability and a negative value indicates an increase in protein stability). (D) The F8 and F305 aromatic side chains (dark violet) form a pi-pi T-stack and F8 in addition interacts with N151, I152, and R298 (green sticks, PDB entry 7ALI (51)). F8 is located within the homodimer interface, with chains A and B colored light violet and blue, respectively. The conjugated p-orbitals of the F305 side chain are located nearby of the I152 and S123 backbone oxygen atoms, potentially leading to electronic repulsion. In the F305L mutant, the distance of the leucine side chain (yellow sticks) to these oxygen atoms is considerably increased. (E) Bar plots showing the percentage of frames against C-term/C-term distance or C-term/P1 distance for WT (F) Bar plots showing the percentage of frames against C-term/C-term distance or C-term/P1 distance for L167F/F305L/F8L mutant. (G) Scatter/cluster plot of WT (orange) and the L167F/F305L/F8L (dark green) mutant C-term-C-term and C-term/P1 distances.