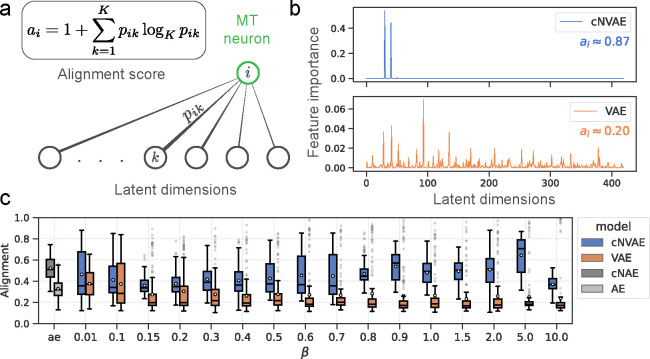
Figure 8:
Hierarchical models (cNVAE, cNAE) are more aligned with MT neurons since they enable sparse latent-to-neuron relationships. (a) Alignment score measures the sparsity of permutation feature importances. when all latents are equally important in predicting neuron ; and, when a single latent predicts the neuron. (b) Feature importances are plotted for an example neuron (same as in Fig. 6b). cNVAE predicts this neuron’s response in a much sparser manner compared to non-hierarchical VAE . Supplementary section 9.5 contains a discussion of our rationale in choosing these values. (c) Alignment across values, and autoencoders (ae).